Unraveling the Secrets of KPC Variants: The Rise of Cefiderocol Resistance!
2025-08-25
Author: Rajesh
Introduction to KPC Variants and Cefiderocol Resistance
In the quest to understand antibiotic resistance, especially concerning cefiderocol, researchers have taken a deep dive into the mutational landscape of KPC (Klebsiella pneumoniae carbapenemase) variants. Recent studies have demonstrated that random mutagenesis can yield a plethora of mutant strains, showcasing the adaptive capabilities of these bacteria under treatment pressure.
The Experiment: From Mutant Libraries to Resistance
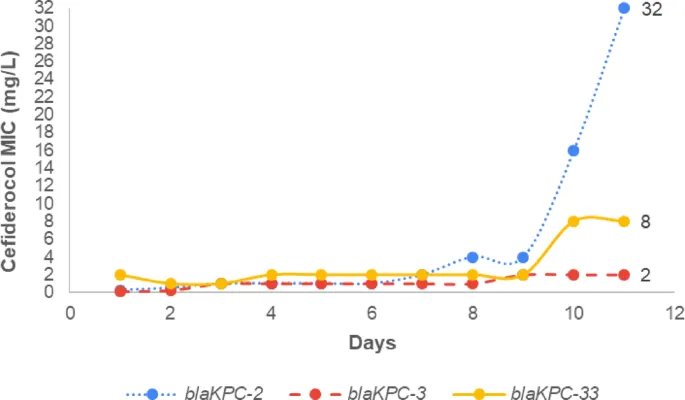
During random mutagenesis, a staggering 130,000 colonies were generated from three KPC variants: blaKPC-2, blaKPC-3, and blaKPC-33. Each colony represents a unique mutation, giving insight into how these variants can evolve resistance to cefiderocol—a crucial weapon against multidrug-resistant infections.
Resistance Mechanisms: The Numbers Speak Volume!
Initial cefiderocol Minimum Inhibitory Concentrations (MICs) for these variants were relatively low. However, after just ten days of exposure, MICs skyrocketed: over 128-fold increase for blaKPC-2, 17-fold for blaKPC-3, and fourfold for blaKPC-33. This rapid evolution highlights a concerning trend in antibiotic resistance.
Phenotypic Traits: Identifying the Resistance Profiles
Antibiotic susceptibility testing unraveled a common resistance profile among the mutants, with all showcasing resistance to ceftazidime and other beta-lactams. Interestingly, these mutants displayed altered susceptibility, becoming more sensitive to carbapenems—an evolutionary trade-off that could complicate treatment options.
Genomic Insights: Sanger Sequencing Reveals Key Mutations
Sanger sequencing identified critical mutations: blaKPC-2 mutants exhibited D179Y and D209V, while blaKPC-3 yielded the L169P mutation. These genetic changes are pivotal as they enhance the enzyme's hydrolytic capability against cefiderocol.
Beyond KPC: The Search for Other Resistance Mechanisms
Whole genome sequencing (WGS) painted a broader picture. Interestingly, no new mutations surfaced in genes associated with porins or transporters. However, all genomes analyzed had insertions in the cirA gene, essential for cefiderocol uptake, further complicating resistance mechanisms.
Intriguing Implications for Clinical Practice
The ability of KPC variants to adapt quickly under cefiderocol exposure is alarming, especially considering clinical relevance. The crossover resistance against ceftazidime implies a potential rapid selection of resistant mutants during treatment, threatening our remaining therapeutic options.
Conclusion: Navigating the Complexities of Antibiotic Resistance
This comprehensive investigation sheds light on how KPC variants interact with cefiderocol and adapt to survive. The findings underscore the urgent need for integrated therapeutic strategies and surveillance systems to curb the emergence of resistance, ultimately preserving the efficacy of crucial antibiotics like cefiderocol.
Call to Action!
As KPC variants continue to evolve, it is imperative for researchers, clinicians, and healthcare systems to collaborate. Understanding these mechanisms isn’t just about science; it’s about ensuring effective treatments for future generations. Let’s act now to combat the growing threat of antibiotic resistance!




 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)