Revolutionary Technique Unlocks Secrets of Bacterial Gene Activity
2025-05-08
Author: Ming
A Game-Changer in Bacterial Research
Imagine being able to analyze the gene activity of every single bacterial cell in a colony in a way that’s quicker and more effective than ever before! Researchers at Julius-Maximilians-Universität Würzburg have developed a groundbreaking technique that does just that.
Understanding Bacterial Diversity Is Key
Bacterial populations are anything but uniform. Within a colony, some bacteria are on the brink of cell division, while others are adapting to new environmental challenges. This intricate diversity is especially crucial when studying pathogens, as understanding these differences can pave the way for better treatment options.
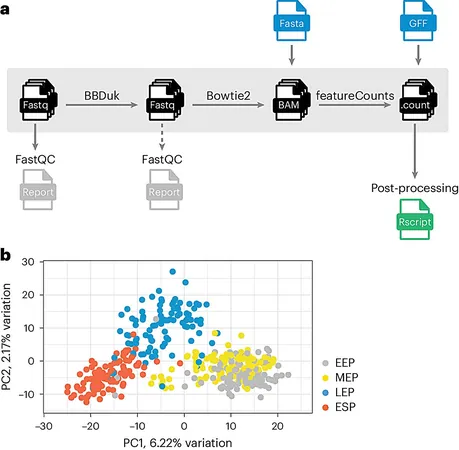
Introducing MATQ-seq
In 2020, a novel variant of single-cell transcriptomics, called MATQ-seq, was introduced by researchers from Würzburg. This ingenious method leverages messenger RNA (mRNA) to pinpoint which genes are active in individual bacterial cells at any given moment, making it a powerful tool for observing how these cells react to antibiotics and other environmental changes.
A Protocol for Success
Recently, the team has fine-tuned this approach, releasing a detailed protocol in the journal Nature Protocols. This step-by-step guide outlines how to create single bacterial transcriptomes using MATQ-seq, complete with detailed experimental and computational analysis.
Achieving Impressive Results
Dr. Christina Homberger, a key researcher in the project, highlighted the protocol's efficiency: "We’ve achieved a staggering 95% cell retention rate!" This means that nearly every cell analyzed ends up contributing to the final gene libraries, a huge improvement over existing methods which can lose up to 70% of samples.
Analyzing Gene Activity Like Never Before
With MATQ-seq, researchers can now reliably measure the activity of between 300 to 600 genes per bacterial cell. According to Dr. Fabian Imdahl, this is significantly more than what other methods can provide. This level of detail allows scientists to easily determine what each bacterium is doing and how it's adapting to its surroundings.
A Unique Collaboration Platform
The impact of this research doesn’t stop here. Professor Jörg Vogel, director of HIRI and IMIB, indicates that this protocol lays the groundwork for creating a first-of-its-kind global platform for microbial single-cell RNA sequencing right in Würzburg. Named the Center for Microbial Single-Cell RNA-seq (MICROSEQ), this facility will enable researchers worldwide to access advanced technologies for analyzing individual bacterial cells.
With this revolutionary approach, the future of bacterial research looks brighter than ever, promising new insights and potential breakthroughs in the understanding and treatment of infectious diseases.




 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)