Unveiling Disease: Revolutionary Framework Sheds Light on Diseased Tissues
2025-04-28
Author: Wei Ling
In the quest to decode what drives disease progression in our bodies, scientists are now armed with a groundbreaking tool that goes beyond simple snapshots of isolated cells. Introducing MESA (Multiomics and Ecological Spatial Analysis)—a cutting-edge computational method exposing the intricate ecology of diseased tissues!
Detailed in a recent Nature Genetics study, this innovative approach emerged from a powerhouse collaboration involving renowned institutions such as MIT, Stanford University, Weill Cornell Medicine, and the Broad Institute. Spearheaded by a team from Stanford, MESA applies an ecological perspective to the analysis of tissue health.
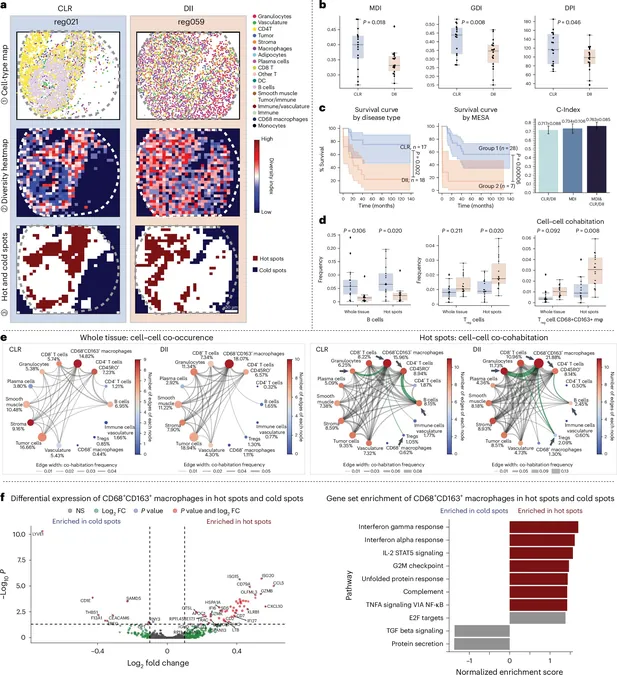
By harnessing spatial omics data—which uncovers not only the identities of various cells, but also their locations within tissue samples—MESA provides a detailed map of tissue "neighborhoods." This tool allows scientists to decode the structural significance of these areas as they relate to disease.
Alex K. Shalek, a key figure in the research and director at MIT's Institute for Medical Engineering and Science, passionately states, "By merging concepts from different fields, we gain insights into how tissues are organized locally and how these arrangements shift in various disease scenarios. This could revolutionize diagnostics and spotlight new avenues for therapies."
Using an ecological lens, postdoc Bokai Zhu explains how MESA likens cell types to species in an ecosystem. "Just as ecologists study how animal species interrelate, we analyze the relationships among T cells and B cells within tissues. This analogy lets us evaluate cellular 'biodiversity' and its fluctuations during disease progression."
For instance, MESA revealed significant findings in liver cancer samples, pinpointing areas where tumor cells consistently cluster with macrophages, indicating they might influence unique disease outcomes.
Zhu adds, "Our method interprets tissues like ecosystems, revealing significant 'hotspots' that could indicate early disease stages or responses to treatment. This insight propels new opportunities for precision medicine."
A standout feature of MESA is its ability to enrich tissue data computationally, minimizing the need for extensive new experiments. By tapping into publicly available datasets, MESA amplifies existing tissue samples with valuable information like gene expression profiles—enhancing our understanding of spatial dynamics, especially when juxtaposing healthy and diseased tissues.
In trials involving diverse datasets and tissue types, MESA has unearthed spatial structures and crucial cell populations that had previously slipped beneath the radar. By integrating various omics data—transcriptomics, proteomics, and beyond—it assembles a richly layered depiction of tissue architecture that could transform research.
Though currently available as a Python package geared toward academic and translational research, MESA's potential is vast yet untapped in routine clinical settings. However, pharmaceutical companies are keenly interested, especially for drug trials where understanding tissue responses is vital.
As Zhu concludes, "This is merely the beginning! MESA opens gateways to applying ecological theories in unraveling the complex spatial dynamics of diseases—ultimately enhancing our ability to predict and treat them more effectively."


 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)