Unlocking the Secrets of Italian Mosquitoes: New DNA Barcoding Techniques for Identifying Native and Alien Species
2024-09-28
Author: Li
Introduction
Mosquitoes, members of the Culicidae family, are infamous worldwide as vectors of deadly diseases that pose a significant risk to human health. With the alarming rise in invasive mosquito species across multiple regions, experts are increasingly concerned about their effects on public health. Traditional identification methods largely rely on the morphological examination of specimens, but this approach has severe limitations, especially concerning the expertise required and its inability to accurately identify damaged samples. Enter DNA barcoding—an innovative molecular technique that transforms mosquito identification and monitoring.
The Study’s Ambitious Goals
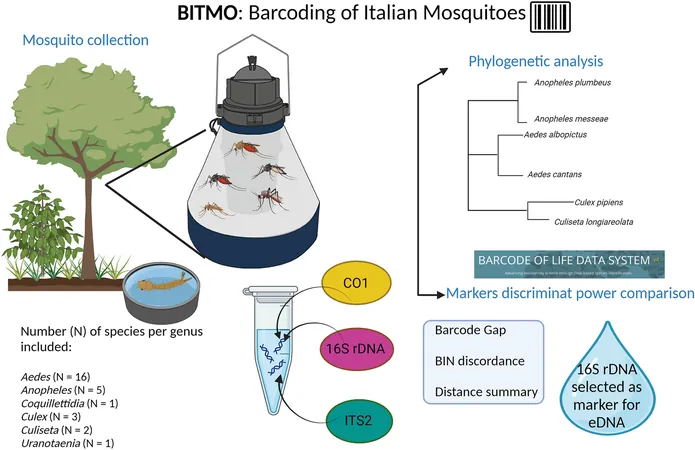
The recent research initiative, titled "Barcoding of ITalian MOsquitoes" (BITMO), aimed to generate and validate a comprehensive DNA barcoding reference library. The primary objectives were threefold: to design universal primers targeting the 16S ribosomal RNA (rRNA) gene, to build a 16S rRNA reference library focusing on Italian mosquito species, and to evaluate the effectiveness of the 16S rRNA gene compared to the widely used cytochrome c oxidase subunit 1 (COI) and internal transcribed spacer 2 (ITS2) markers.
Methodology Overview
This study analyzed a total of 28 mosquito species spanning six different genera: Aedes, Anopheles, Coquillettidia, Culex, Culiseta, and Uranotaenia. DNA was extracted from whole mosquito bodies, which led to the successful generation of DNA sequences through Sanger sequencing. The findings were meticulously analyzed using the Barcode of Life Data Systems (BOLD), and phylogenetic analyses were conducted to clarify relationships among species.
Key Findings
The researchers generated new sequences for the 16S rRNA, COI, and ITS2 genes. Notably, the 16S rRNA gene emerged as a vital marker capable of accurately distinguishing mosquito species, possessing a discriminatory capacity comparable to that of COI. Furthermore, there are certain advantages when using the 16S rRNA gene in eDNA applications due to the shorter size of the DNA fragments, making it ideal for environmental samples where DNA degradation likely occurs.
Italy’s Mosquito Landscape
Italy boasts a diverse mosquito fauna composed of 65 species from eight genera, including the notorious tiger mosquito Aedes albopictus which has expanded its range from Asia. In recent years, species such as Aedes koreicus and Aedes japonicus have also been making their presence felt, prompting fears about their potential as vectors for tropical diseases. The historical absence of certain species, including Aedes aegypti, which hasn’t been recorded in Italy since 1971, raises red flags about the dynamic nature of mosquito populations.
The Urgent Need for Monitoring
The rise of invasive mosquito species (IMS) corresponds with increasing reports of locally transmitted diseases in Italy, including chikungunya and dengue fever. These developments underline the critical need for accurate entomological monitoring, which can no longer solely rely on traditional methods. There is a pressing demand for molecular-based taxonomic approaches to ensure effective identification, particularly in environments abundant with insect species.
A Future with Enhanced Monitorization
As the BITMO project successfully compiled a robust reference library for Italian mosquitoes, the added sequences will significantly bolster research into biodiversity, conservation efforts, and monitoring initiatives concerning mosquito populations. Emphasizing molecular identification methods paves the way for more accessible and reliable monitoring of mosquitoes, promising a proactive approach to controlling these significant public health threats.
Conclusion
In conclusion, this groundbreaking research has provided essential insights and tools for the future of mosquito monitoring in Italy. With innovative techniques and comprehensive DNA libraries, scientists are better equipped to tackle the challenges posed by these pestilent pathogens. Stay tuned—will we find more invasive species that challenge our health systems? Or will the new DNA tools help us reclaim control over these tiny terrorizers? Only time will tell!



 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)