Unlocking the Future of Macadamia: Genome Sequencing of All Four Species Reveals Game-Changing Insights!
2024-11-04
Author: Jia
Introduction
In a groundbreaking development for agriculture, a dedicated research team has successfully sequenced the genomes of all four Macadamia species, opening the door to revolutionary crop improvement strategies for this lucrative nut. This monumental achievement not only sheds light on essential genetic traits but also aims at enhancing disease resistance, climate adaptability, and yield—the vital components needed to advance macadamia cultivation and meet global demands.
The Macadamia Genus
The Macadamia genus, which originally hails from the lush landscapes of eastern Australia, comprises four distinct species: Macadamia integrifolia, M. tetraphylla, M. ternifolia, and M. jansenii. However, commercially viable cultivation primarily revolves around the first two species and their hybrids, limiting the genetic diversity of this sought-after crop. This research aspires to rectify this issue by expanding the genetic resources available through comprehensive genome assemblies.
Current Production Challenges
Current macadamia production is heavily reliant on a constricted genetic base, with Hawaiian germplasm dominating the landscape. This narrow foundation has hindered genetic diversity, leaving crops vulnerable to disease and climate variability. The study, published in the journal Tropical Plants on October 21, 2024, is set to broaden the gene pool, ensuring enhanced resilience while preserving the high-quality traits that consumers love, such as flavor richness and oil content.
Advanced Sequencing and Results
Utilizing the cutting-edge PacBio HiFi long-read sequencing technology, researchers achieved remarkable results, with sequencing depths ranging from 27X to 42X. The genome assemblies yielded highly contiguous structures with N50 values surpassing 45 Mb, signifying outstanding genome completeness—a leap forward in genetic studies.
Species Insights and Findings
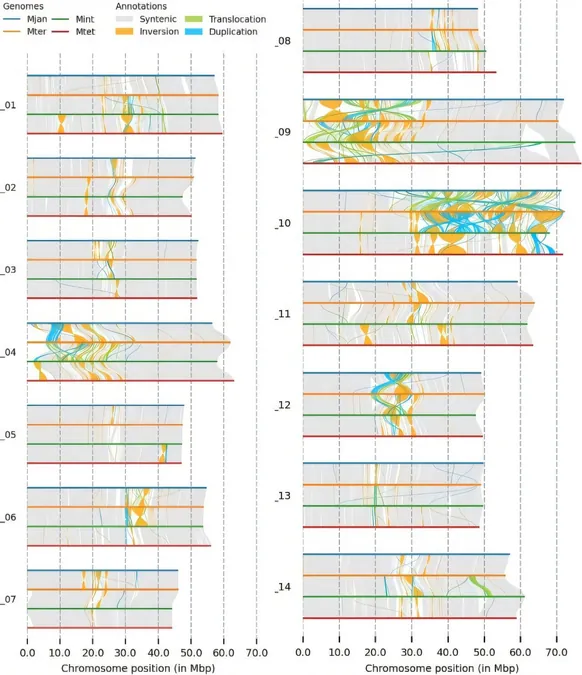
Among the species, M. integrifolia showcased the highest number of contigs, while M. tetraphylla demonstrated the lowest. A BUSCO analysis later affirmed the genome completeness rate of over 97%, confirming the majority of the genes as single-copy. The investigation also revealed chromosome-level assemblies, stating genome sizes between 735 Mb and 795 Mb, with M. tetraphylla showcasing the most extensive collapsed assembly and M. jansenii the least.
Structural Variations and Gene Analysis
With structural variations noted on chromosomes 9 and 10, the researchers delved deeper into genome annotation. Astonishingly, 61% to 62% of the genomes were found to consist of repetitive elements, highlighting the complexity of the genetic landscape. The predicted gene count ranged from 37,198 to 40,534, with crucial genes for fatty acid biosynthesis and antimicrobial properties being conserved across the species.
Implications and Future Prospects
This pivotal research lays a robust foundation for refining macadamia breeding programs, utilizing the genetic diversity inherent in wild species to tackle pressing challenges: advancing disease resistance and bolstering climate resilience.
Dr. Robert J. Henry, the leading researcher, elaborates on the implications, stating, "Our access to the genomes of all four macadamia species opens unprecedented avenues for enhancing crop resilience and productivity. This comprehensive genomic data forms the bedrock for informed breeding programs essential to confronting the demands of our evolving climate and expanding global markets."
Conclusion
As the macadamia industry prepares for a future where resilience and productivity are key, this comprehensive sequencing study promises to usher in significant advancements in breeding methodologies. With global production set to soar, the results represent a critical tool for breeders and researchers committed to safeguarding the future of this economically vital crop in an increasingly unpredictable world.


 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)