Uncovering Hidden Threats: Shotgun Metagenomic Sequencing Reveals Pathogens in HIV-Infected Children Suffering from Non-Malarial Febrile Illnesses
2025-01-21
Author: Li
Introduction
Non-malarial febrile illnesses (NMFI) present a formidable challenge, particularly among HIV-infected children in Uganda, leading to severe health complications and heightened morbidity. While conventional diagnostic methods predominantly focus on specific pathogens, revolutionary techniques like shotgun metagenomic sequencing pave the way for a richer, more comprehensive understanding of the gut microbiome and its role in NMFI management.
Research Methods
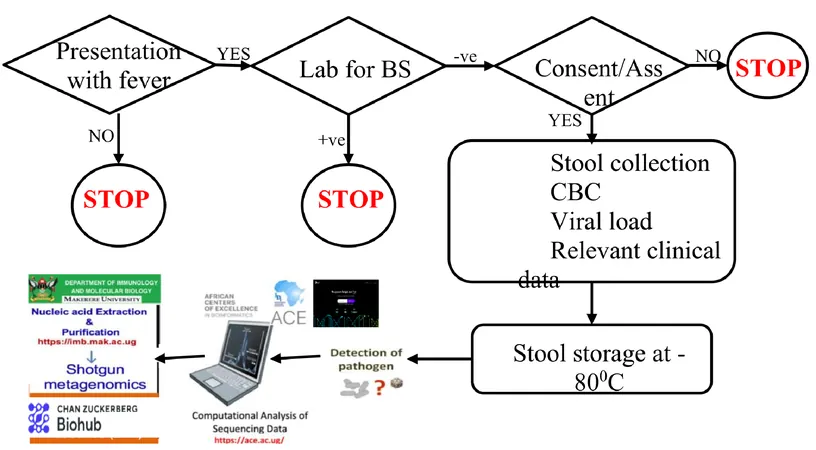
This groundbreaking study was conducted at Baylor Children’s Clinic Uganda, where researchers utilized shotgun metagenomic sequencing to analyze stool samples from HIV-positive children diagnosed with NMFI. Over 140 stool samples were carefully collected, and advanced DNA extraction methods were employed at Makerere University’s Molecular and Genomics Laboratory. The extracted DNA was sent to the Chan Zuckerberg Biohub in San Francisco for in-depth shotgun metagenomic sequencing, allowing researchers to delve deep into the microbial ecosystems contributing to febrile illnesses.
Key Findings
The study unearthed an astonishingly vast array of microbial taxa in the analyzed stool samples. Among the discovered potentially pathogenic microbes were Trichomonas vaginalis, Candida albicans, Giardia, and Bacteroides. These findings highlighted the intricate web of microbial interactions that may exacerbate NMFI's clinical impacts in HIV-infected children. Taxonomic profiling not only identified well-known pathogens but also pointed towards potential new links with NMFI, shedding light on the underlying mechanisms of febrile illnesses in this vulnerable demographic.
In the greater context of Uganda, where malarial infections have historically overshadowed other febrile conditions, the study underlines the critical need for improved diagnostic practices that go beyond traditional methods. With 284 per 1000 individuals at risk of malaria between 2020 and 2021, relying solely on specific pathogen detection leaves many infections undiagnosed. Comprehensively tackling NMFI requires innovative surveillance strategies to cover the diverse range of responsible pathogens.
Conclusion and Implications
This study showcased the immense potential of shotgun metagenomics as a novel avenue to understand the complexities of NMFI in children living with HIV. By uncovering a broader spectrum of pathogens, the technique not only enhances our understanding of infectious diseases in vulnerable populations but paves the path towards more targeted therapeutic approaches.
It is clear that the integration of metagenomic techniques in clinical diagnostics could revolutionize patient care, allowing for individualized treatment plans that better address the diverse etiologies of fever in HIV-infected children. Moving forward, comprehensive healthcare programs need to emphasize early detection and the development of extensive diagnostic tools that address the dynamic epidemiology of infections in this demographic.
In conclusion, as the fight against HIV and the associated complications continues, leveraging modern sequencing technologies will be crucial in improving clinical outcomes for affected children in Uganda and beyond. The results of this research not only serve as a beacon for future studies but also as a clarion call for immediate action in public health initiatives to combat the neglected burden of non-malarial febrile illnesses.
Call to Action
Stay informed and support ongoing research in infectious diseases to help protect vulnerable populations like HIV-infected children – their health is our collective responsibility!

 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)