Scientists Unveil the Secrets of tRNA Processing: A Dual-End Cleavage Breakthrough!
2025-07-07
Author: Nur
Unlocking the Blueprint of Protein Synthesis
In the intricate world of cellular biology, transfer RNA (tRNA) plays a crucial role as the essential courier in protein synthesis. These remarkable molecules interpret genetic messages from messenger RNA (mRNA) and deliver the corresponding amino acids to ribosomes—our cells’ protein factories. But before tRNA can spring into action, it requires precise trimming and modification.
A Groundbreaking Discovery by Kyushu University Researchers
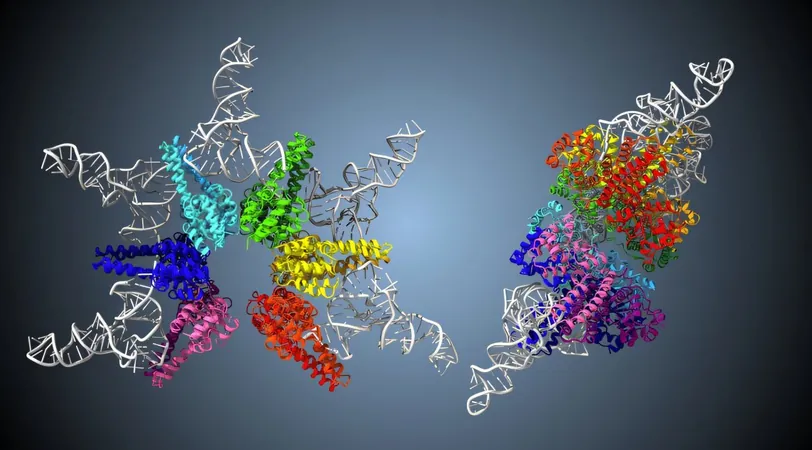
A team from Kyushu University has made a significant discovery: they have uncovered the mechanism behind the dual-end cleavage of tRNA, facilitated by the smallest known protein-based tRNA-processing enzyme named HARP. This enzyme forms a unique star-shaped assembly of twelve protein molecules, enabling it to perform critical cuts at both the 5' and 3' ends of tRNA. Their findings, published in *Nature Communications*, promise to have wide-ranging implications for synthetic biology and biotechnology.
The Complexity of tRNA Processing
In cellular systems, proteins must often undergo various modifications to become functional. For tRNA, this involves snipping off excess segments from its ends, specifically its 5-prime (5') and 3-prime (3') ends. The well-known enzyme RNase P, found across nearly all forms of life, is primarily responsible for this trimming process.
Meet HARP: The Tiny but Mighty Enzyme
HARP, short for Homologs of Aquifex RNase P36, is notable for its small size and unique six-pointed star structure. Until now, the specifics of how HARP executes these intricate tasks had remained a mystery. Professor Yoshimitsu Kakuta, who led the study, emphasized the use of advanced cryogenic electron microscopy (cryo-EM) to visualize HARP interacting with pre-tRNA and reveal its processing methods.
Fascinating Findings on tRNA Cleavage
Using cryo-EM, researchers discovered that HARP binds pre-tRNA molecules on five distinct sites, forming a radial structure. This star-shaped enzyme acts almost like a "molecular ruler," measuring distances to pinpoint the precise cleavage site. Intriguingly, this sophisticated mechanism has been observed in other RNase P enzymes as well, suggesting a remarkable instance of convergent evolution.
Unexpected Revelations from the Research
The team also made a surprising discovery: HARP binds to only five pre-tRNA molecules instead of the ten previously expected, leaving seven active sites vacant. Further tests revealed that these vacant sites are actually used for cleaving the 3' end of pre-tRNA, showcasing HARP’s bifunctionality.
An Evolutionary Triumph in tRNA Processing
According to Kakuta, the multi-functional abilities of HARP represent an evolutionary strategy, enabling organisms with compact genomes to achieve complex tasks with fewer structural components. This groundbreaking research could pave the way for the next generation of synthetic biology tools, offering exciting prospects for innovation in biochemistry.
A New Era of Synthetic Biology Awaits!
With discoveries like these, the potential for designing artificial enzymes and RNA processing systems is limitless! As scientists continue to delve into the mechanics of tRNA and other essential cellular processes, we stand at the brink of a revolution in how we understand and manipulate biological systems.


 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)