Revolutionizing Species Tree Inference: Meet ROADIES!
2025-05-05
Author: Yu
A Game-Changer for Evolutionary Research
At the University of California San Diego, a groundbreaking team of engineers is changing the way researchers interpret the evolutionary relationships among species. Their innovative tool, ROADIES, empowers scientists, regardless of their expertise level, to construct detailed species trees directly from raw genome data with minimal computational resources.
Why Species Trees Matter
Understanding species trees is crucial not just for tracing evolutionary history but also for significant applications in medicine and conservation. These trees can pinpoint functional genomic regions for potential drug targets, correlate physical traits with genetic changes, anticipate zoonotic disease outbreaks, and inform conservation strategies.
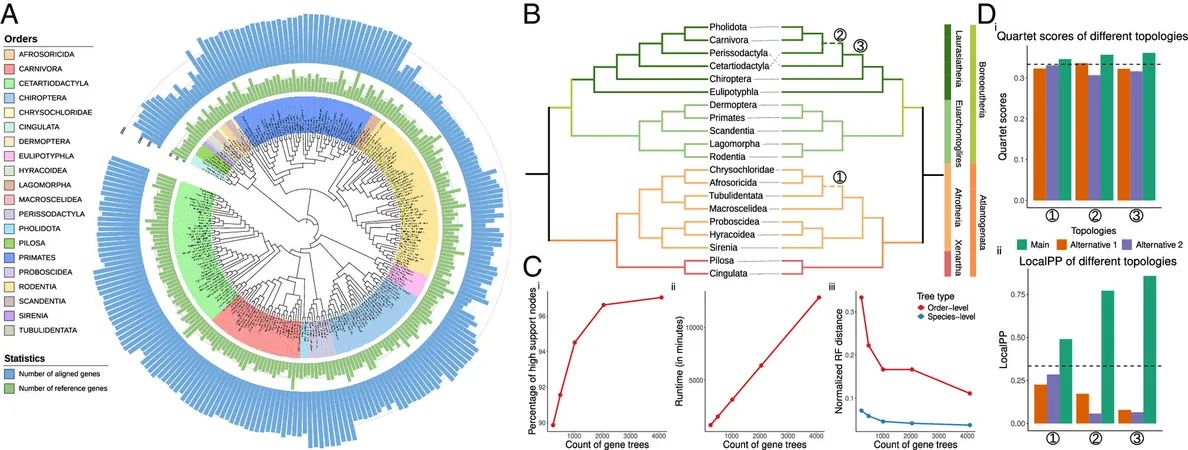
ROADIES: Fast, Accurate, and Easy to Use
In a recent publication in the *Proceedings of the National Academy of Sciences*, the team's lead researcher, Yatish Turakhia, demonstrated that ROADIES can produce species trees with comparable quality to state-of-the-art methods, but with far less time and effort involved. The study included an impressive range of species - placental mammals, pomace flies, birds, and budding yeasts - showing the tool's versatility.
Anshu Gupta, the study's first author and a Ph.D. student at the Jacobs School of Engineering, noted, "While advances in sequencing technology have made genome assembly more accessible, accurately constructing species trees remains a challenge for many researchers."
Introducing a Fully Automated Process
ROADIES—which stands for Reference-free, Orthology-free, Annotation-free, Discordance-aware Estimation of Species Trees—breaks new ground in phylogenetics with its fully automated workflow that guarantees precision.
Instead of relying on pre-defined genomic regions, which often complicate the process, ROADIES utilizes a random sampling approach across genomes. This approach allows researchers to skip the usually necessary genome annotation, achieving impressive accuracy from seemingly random loci.
Turakhia remarked, "It may be counterintuitive, but our findings show that reconstructing species trees from these random samples is not only effective but might also provide unique advantages in aligning with evolutionary models."
Innovation in Automation
Another remarkable feature of ROADIES is its ability to incorporate gene copies that exist multiple times within genomes, a common scenario across many species. This capability comes from integrating advanced techniques from UC San Diego, enabling the tool to bypass traditional orthology inference.
By eliminating the need for genome annotation and orthology inference, ROADIES simplifies the process and drastically reduces computing power requirements. The study confirms its scalability to datasets involving hundreds of genomes, producing phylogenies consistent with extensive expert-led projects, yet requiring a fraction of the effort.
Preparing for the Future of Genomic Research
The research team is dedicated to enhancing ROADIES further, targeting improvements like the integration of new species into existing trees and potentially leveraging GPUs to analyze tens of thousands of genomes.
Turakhia emphasized the urgency, stating, "Major initiatives are already in the works to sequence thousands of species, possibly all existing eukaryotic species on Earth. We aim to ensure ROADIES is equipped to handle that monumental challenge!"


 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)