Revolutionizing Medical Imaging: AI Transforms Mass Spectrometry into Virtual Histology
2025-08-04
Author: Mei
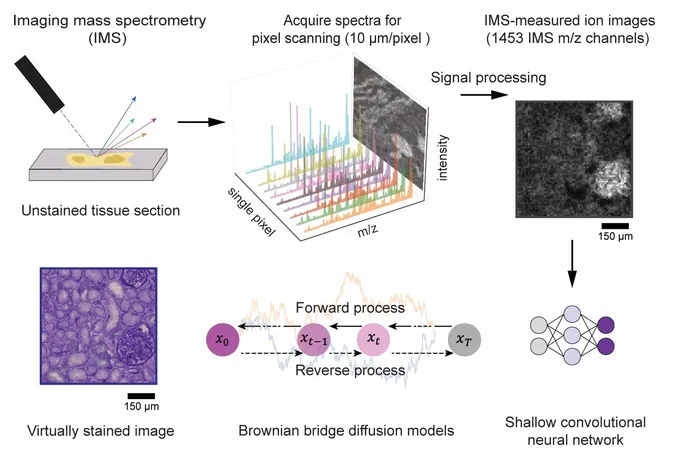
In a groundbreaking development, an international research team from UCLA, Vanderbilt University, and Delft University of Technology has harnessed artificial intelligence (AI) to advance imaging mass spectrometry (IMS) by enabling virtual histological staining. Their innovative work has been featured in the journal Science Advances.
This collaboration marks a significant leap forward, enhancing spatial resolution and cellular detail without the need for traditional chemical staining. By employing a cutting-edge diffusion-based generative model, the researchers have digitally rendered images that rival the quality of conventional histochemical stained samples, all while preserving precious tissue specimens.
Imaging mass spectrometry is renowned for its ability to map a multitude of molecular species in biological tissues with remarkable specificity. However, traditional IMS methods often struggle with low spatial resolution and insufficient cellular detail, key elements necessary for accurately interpreting molecular profiles in relation to tissue architecture.
To tackle these limitations, the team introduced an innovative diffusion-based virtual staining approach. This technique converts low-resolution, label-free IMS data into high-resolution brightfield microscopy images that closely mimic histochemically stained samples, particularly those stained with Periodic Acid–Schiff (PAS). PAS staining is crucial as it highlights various essential components like polysaccharides and glycoproteins in tissues.
Notably, this AI framework achieves high fidelity despite IMS data having pixel sizes nearly ten times larger than standard optical microscopy images. Professor Aydogan Ozcan from UCLA remarked, "This diffusion-based approach revolutionizes the interpretability of mass spectrometry images, linking molecular specificity with cellular structure—all without chemical tissue stains."
In blind tests conducted on human kidney tissues, the virtually stained images demonstrated a striking resemblance to chemically stained samples. This breakthrough allows pathologists to pinpoint critical renal structures and disease indicators directly from the virtual images.
Additionally, the researchers fine-tuned the noise sampling process during AI inference to ensure consistently reliable staining results, paving the way for broader applications in both clinical and research settings.
This technique heralds transformative potential for IMS-driven biomedical research and diagnostics, eliminating the cumbersome chemical staining processes and complex image alignment tasks. Not only does it safeguard tissue integrity for further molecular analyses, but it also accelerates mass spectrometry-based workflows in molecular histology.
Looking ahead, Professor Ozcan expressed optimism, stating, "We envision this approach will usher in new avenues in spatial biology and clinical diagnostics. By creating high-quality histological images directly from mass spectrometry data, we can significantly streamline workflows and enhance biomedical discoveries."

 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)