Revolutionary Python Tool Transforms Single-Cell Lineage Tracing and Speeds Up Cancer Research
2025-07-25
Author: Sarah
Introducing scLT-kit: The Future of Cell Analysis
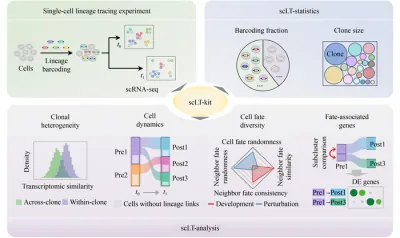
Researchers at Tsinghua University have unveiled an innovative Python toolkit known as scLT-kit, designed to automate the analysis of single-cell lineage tracing data. Imagine it as a high-tech GPS for understanding cell ancestry—this tool simplifies complex, high-dimensional data, allowing scientists to easily track the developmental pathways of individual cells and their responses to treatments.
A Game Changer for Medicine and Cancer Research
The ability to decipher how cells evolve is vital in fields like regenerative medicine and oncology. With scLT-kit, lineage tracing becomes accessible and efficient, expediting investigations into tissue development, drug resistance, and disease progression. This advancement is set to empower biologists, pharmaceutical experts, and health policymakers as they assess cellular responses to therapies, potentially leading to groundbreaking diagnostic and treatment strategies.
Streamlining Complex Analyses with Ease
"We’ve transformed a previously tedious analysis process into a smooth and reproducible workflow with scLT-kit," says Professor Jin Gu. "Our aim is to enable every lab—be it studying development, drug resistance, or disease progression—to derive lineage insights from single-cell data in mere minutes instead of months."
Revolutionary Insights into Cell Behavior
The researchers' analyses revealed astounding findings with scLT-kit: - It efficiently processes time-series single-cell RNA sequencing data embedded with heritable barcodes, offering swift quality assessments and statistics on barcoding effectiveness and clone sizes. - In experiments involving developmental datasets, such as blood progenitors and C. elegans embryos, cells sharing identical barcodes exhibited striking similarities in gene expression. - In contrast, tumor cells treated with EGFR inhibitors showed diminished similarity within clones, indicating the high diversity of surviving cancer cells. - The toolkit quantifies cell fate variability, showcasing that normal cell development follows more predictable paths, while drug-treated cancer cells display more randomness in their fate choices.
An All-in-One Python Solution for Researchers
scLT-kit integrates standard single-cell RNA-seq processing with two dedicated modules focused on lineage data. The scLT-statistics module assesses barcoding fractions and clone sizes at various time points, while the scLT-analysis module constructs lineage networks to visualize cellular transitions over time through interactive Sankey plots. Additionally, it features four quantitative measures to evaluate cell fate randomness and neighborhood similarity, linking gene expression changes to specific fate decisions—all packaged into an easy-to-install toolkit available on PyPI and GitHub.
Lowering Barriers in Single-Cell Research
By offering robust, automated workflows, scLT-kit lowers entry barriers for labs exploring single-cell lineage tracing during both development and disease conditions. With its combination of quality checks, dynamic analysis, and gene insights, this toolkit promises to enrich our understanding of cellular fate decisions, paving the way for advances in medicine and cancer therapy.



 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)