Revolutionary Metagenomic Profiling Method Offers Greater Precision and Efficiency Using Cutting-Edge Technology!
2024-11-13
Author: Jia
Introduction
A groundbreaking advancement in metagenomics has emerged from the collaborative efforts of researchers at Carnegie Mellon University and the University of Toronto. They have unveiled a pioneering k-mer sketching metagenomic profiler known as Sylph, which significantly enhances the speed and accuracy of genomic data analysis, setting a new standard for the scientific community.
Publication and Implications
This innovative method has been documented in detail in the latest edition of Nature Biotechnology, bringing anticipation within the research sector. According to Yun William Yu, an assistant professor at CMU's Ray and Stephanie Lane Computational Biology Department, the rapid advancement in sequencing technology presents both opportunities and challenges. “Sequencing is getting better, which is great because it means we have more data to work with,” he noted. However, the surge in data can lead to prolonged analysis times, which Sylph seeks to mitigate.
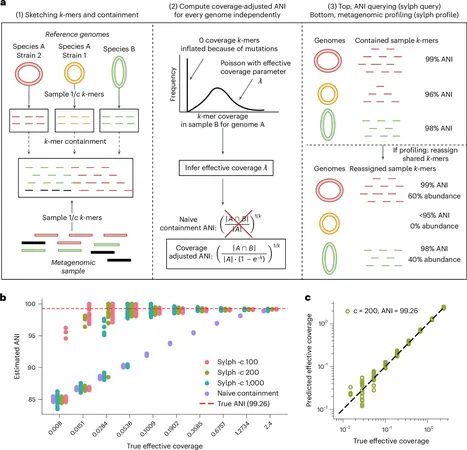
How Sylph Works
Traditional techniques often involve reading sequenced genomic data, matching it to known bacteria like E. coli and C. difficile, and quantifying their presence in a sample, such as that derived from the human gut. Sylph takes an innovative approach by reversing this process. It utilizes known bacterial genomes, breaking them into smaller segments known as k-mers, and compares these segments against the sample. If a specific k-mer is detected in the sample, it indicates the presence of the corresponding bacterium, thereby streamlining the identification process.
Performance Advantages
What sets Sylph apart from its predecessors is its efficient use of computing resources and its ability to process data at unprecedented speeds. Yu, alongside collaborator Jim Shaw, a postdoctoral fellow at Harvard Medical School and Dana-Farber Cancer Institute, has revealed that Sylph maintains its performance advantages even as the datasets grow larger, a crucial factor in the age of expanding sequencing technologies.
Enhancing Computational Workflows
"Sylph enhances computational workflows for complex biological problems, enabling researchers to handle massive datasets effectively," Yu explained. He remarked that while all analytic tools face limitations as data volumes increase, Sylph manages to retain its efficiency longer than existing profiling methods.
Precision and Statistical Models
Moreover, precision is a hallmark of Sylph's design. The tool employs a sophisticated statistical model, grounded in zero-inflated Poisson statistics, to accurately assess the average nucleotide identity (ANI) under low coverage conditions. This is particularly useful when working with low-abundance genomes, allowing researchers to uncover rare genetic information that would otherwise go undetected.
Future Research and Impact
In light of these advancements, Yu is eager to further refine Sylph and incorporate its methodologies into his future research endeavors. With the promise of more precise and resource-efficient genomic analysis, Sylph is poised to make waves in biological research, ultimately contributing to breakthroughs in medicine, ecology, and beyond.
Conclusion
Stay tuned for more developments in this exciting field where technology meets biology in unprecedented ways!



 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)