Revolutionary Deep Learning Method Unlocks Secrets of Protein Conformational Changes
2025-05-15
Author: Sarah
Unveiling the Proteins: A Breakthrough in Biochemical Research
In a groundbreaking study featured in *Nature Communications*, researchers from the University of Wisconsin–Madison have developed a cutting-edge deep learning technique that can automatically identify transition states in protein conformational changes—an essential mechanism behind numerous biological processes.
This innovative tool promises to transform the landscape of biomolecular dynamics research, potentially opening doors for advancements in drug design, biomolecular engineering, and the field of materials science.
A Quest for the "Holy Grail" of Chemistry
For a long time, pinpointing transition states has represented the "holy grail" of chemistry. Unlike standard chemical reactions, protein conformational changes—such as folding and binding—navigate through a maze of multiple metastable intermediate states, leading to a complex web of transition states at free energy barriers.
Until now, research tools have only managed to identify transition states between pairs of metastable configurations, leaving the simultaneous discovery of all transition states in complex biomolecular processes a daunting challenge.
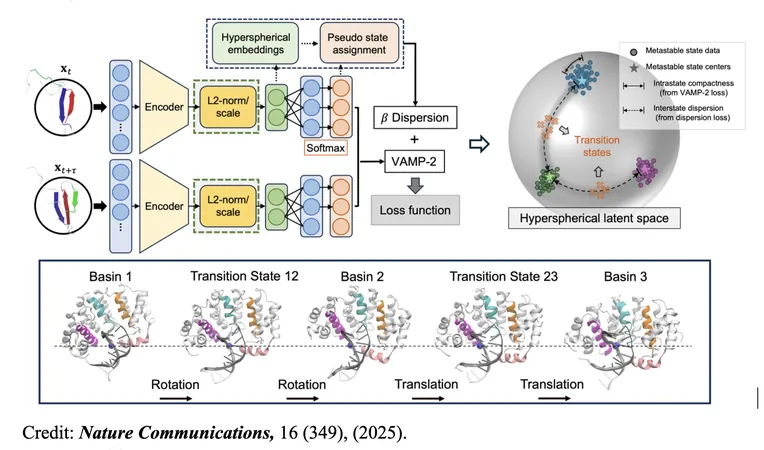
Introducing TS-DAR: A Game-Changing Technique
Meet TS-DAR (Transition State identification via Dispersion and vAriational principle Regularized neural networks), the revolutionary method that breaks through these barriers. TS-DAR employs a deep learning framework inspired by out-of-distribution (OOD) detection—a technique from artificial intelligence designed to recognize data that strays from established patterns.
What makes TS-DAR exceptional is its novel approach to viewing transition states as OOD data—unique structures found at free energy barriers between metastable conformations. By embedding molecular dynamics (MD) data into a hyperspherical latent space, this method efficiently identifies and separates these rare transition states.
A Comprehensive Solution for Protein Dynamics
Providing an all-in-one pipeline for exploring protein dynamics, TS-DAR reveals every transition state involved in biomolecular processes. It’s a significant leap that could reshape our understanding of how proteins function.
"Identifying transition states is one of the most challenging and important tasks in studying protein dynamics," said Prof. Xuhui Huang. "TS-DAR is the first method capable of automatically capturing all transition states at once from MD data, which deepens our understanding of the molecular processes at play."
Proving Its Worth: Testing TS-DAR in Real-World Systems
The research team validated TS-DAR across various systems, including the translocation of a DNA motor protein (AlkD) along a DNA strand. In every instance, TS-DAR surpassed traditional methods in terms of both accuracy and efficiency.
Remarkably, in the AlkD system, the technique provided new insights into the significance of protein-DNA hydrogen bonds, which are crucial for determining the rate-limiting step of AlkD's translocation—an essential mechanism in DNA repair.
Promising Future Ahead: The Broader Implications of TS-DAR
With its capability to detect transition states in intricate biomolecular systems, TS-DAR marks a major advance in molecular dynamics research. This framework’s potential to reliably model highly dynamic processes could pave the way for the creation of generative AI models, revolutionizing the prediction and manipulation of biomolecular dynamics.
 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)