Revolutionary Database Unveils Secrets of Protein Behavior Under Stress
2025-07-11
Author: Siti
Unlocking the Mysteries of Protein Localization
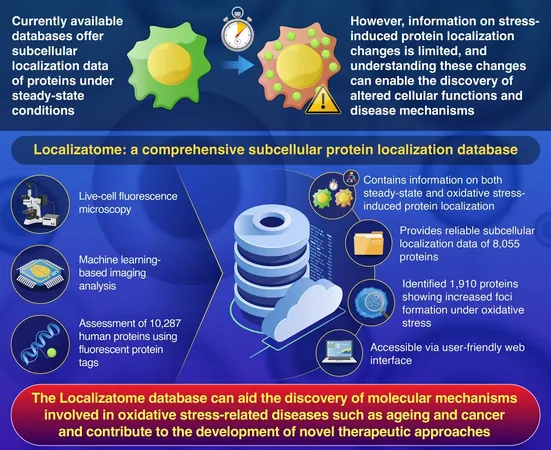
A groundbreaking study from Japan has unveiled a novel, high-throughput fluorescence microscopy system, combined with advanced machine learning algorithms, to track oxidative stress-related changes in how proteins are localized within cells. Researchers have crafted an extensive resource dubbed the Localizatome, cataloging the subcellular localization data of an astounding 10,287 human proteins.
What is Localizatome?
Localizatome isn't just any ordinary database; it provides a dual insight into the stable subcellular localization of proteins and the dynamic shifts that take place when cells experience oxidative stress. Published in the prestigious journal Database, this work paves the way for a deeper understanding of cellular mechanisms related to health disorders and therapeutic strategies.
The Importance of Protein Localization
Proteins are essential biological macromolecules that regulate a variety of functions across cells, tissues, and organ systems. Their effectiveness hinges significantly on being in the right place, at the right time, and in the right amounts within the cell—a concept known as subcellular localization. While previous databases have offered static localization data, they often fall short in detailing how proteins react dynamically under stress conditions.
A Collaborative Effort
The Localizatome project was spearheaded by Professor Hiroshi Asahara from the Department of Systems BioMedicine at Science Tokyo, in collaboration with experts from Osaka University, RIKEN, Musashino University, and the National Institute of Advanced Industrial Science and Technology. Asahara notes that oxidative stress, characterized by the overproduction of reactive oxygen species, significantly contributes to the onset and progression of diseases such as cancer and aging.
"Understanding how proteins behave under oxidative stress is crucial to unraveling the mechanisms behind these diseases," Asahara explains. Localizatome captures the necessary changes in protein localization resulting from such stress.
Innovative Techniques at Work
The research utilized HeLa cells—human cells modified to express 10,287 human proteins tagged with fluorescent labels. A custom-built high-throughput microscopy system, featuring a plate transport robot, allowed the team to conduct live-cell fluorescence imaging, tracking localization patterns pre- and post-oxidative stress exposure. Machine learning techniques were then employed to analyze these images to detect variations in protein localization accurately.
Key Findings and Insights
As a result of their meticulous work, the Localizatome database currently features localization data for 8,055 human proteins. Remarkably, they recorded that 1,910 proteins demonstrated unique foci formation patterns when subjected to oxidative stress. Further evaluations linked these proteins to critical biological pathways, including the Hippo signaling pathway, cell division, and protein degradation.
An Accessible Resource for Future Research
Professor Asahara emphasizes the user-friendly nature of the Localizatome database, which allows easy access to fluorescence image data and detailed protein localization information. "This is a significant step towards openness in scientific research, providing invaluable resources to further investigate protein behavior in stressed cellular environments."




 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)