Revolutionary Computational Tool Unveils Secrets of DNA Regulation in Cancer and Genome Editing
2025-04-29
Author: Siti
Unlocking the Mysteries of DNA with KMAP
A groundbreaking collaboration between researchers from the University of Eastern Finland, Aalto University, and the University of Oulu has led to the creation of KMAP—a powerful new computational tool designed to decode DNA sequence patterns with unprecedented clarity. This innovative method, recently featured in Genome Research, offers a cutting-edge way to visualize short DNA sequences, shedding light on the complex behavior of regulatory elements across various biological contexts.
Transforming DNA Analysis Through Visualization
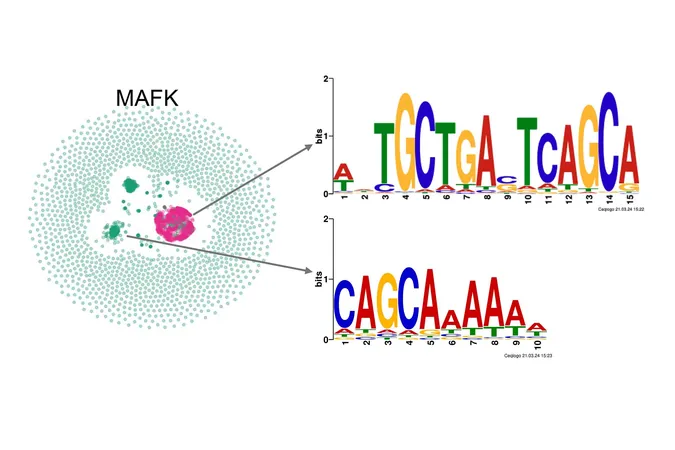
KMAP transforms DNA sequences, known as k-mers, into an easily interpretable two-dimensional format. This visualization allows scientists to swiftly identify and analyze biologically significant DNA motifs. In a revealing re-analysis of Ewing sarcoma data, the researchers employed KMAP to scrutinize genomic areas influential in gene regulation.
Oncogenes and Regulatory Dynamics
Their findings uncovered that the transcription factors BACH1, OTX2, and KCNH2/ERG1 experienced suppression by the oncogene ETV6, only to become active at promoter and enhancer regions once ETV6 was eliminated. Significantly, the team also discovered a previously unidentified DNA motif, CCCAGGCTGGAGTGC, which frequently clustered with BACH1 and OTX2 in enhancer regions. This spatial arrangement hints at a novel regulatory element integral to cancer biology.
Enhancing Genome Editing with KMAP
Furthermore, KMAP played a crucial role in analyzing results from a genome editing experiment utilizing the renowned CRISPR-Cas9 technique at the AAVS1 locus in the human genome. After the CRISPR intervention, researchers observed how cells naturally repair broken DNA using various methods.
Revealing Repair Pathways for Precision Medicine
By visualizing thousands of DNA sequences resulting from this editing process, KMAP pinpointed four prevalent repair patterns—each corresponding to a unique cellular repair pathway. This understanding paves the way for researchers to construct more accurate gene-editing strategies and anticipate the kinds of edits that are most likely to transpire.
A Game-Changer for Molecular Biology
"KMAP provides an intuitive approach for investigating motifs within DNA sequence data," states Dr. Lu Cheng, the lead author of the study. "By mapping the distribution of these short sequences, we enhance our ability to interpret regulatory patterns and uncover their transformations across different biological scenarios."
Professor Gonghong Wei from the University of Oulu adds, "KMAP is versatile and applicable to various sequencing data. Its implications in cancer research for identifying regulatory elements from ChIP-seq data are immense, and it also shows promise for studying RNA-binding proteins and their preferences. The capacity to elucidate structure in complex sequence data positions KMAP as an invaluable tool in molecular biology."


 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)