Revolutionary AI GPS for Microorganisms: A Game Changer for Forensics and Health
2024-11-07
Author: Mei
A groundbreaking research team from Lund University in Sweden has unveiled a cutting-edge AI tool that could forever change the landscape of forensic investigations and public health tracking. This innovative system functions similarly to a GPS navigation service, but instead of helping you find your way to your hotel, it traces the geographical origins of specific microorganisms.
Imagine being able to determine whether someone has recently visited a beach, disembarked from a train in a bustling city center, or taken a serene stroll through a forest—simply by analyzing bacteria. This novel capability has profound implications not only for forensics but also for medicine and epidemiology.
Microorganisms, which include bacteria, are often invisible to the naked eye, yet they hold vast reservoirs of information about the environments they inhabit. The term "microbiome" refers to the collective of microorganisms present in a particular setting, highlighting their complexity and diversity. Historically, determining the geographical origins of microbiome samples has posed significant challenges, but that is about to change.
The research group's recent publication in the journal Genome Biology and Evolution introduces a remarkable tool known as Microbiome Geographic Population Structure (mGPS). This sophisticated instrument harnesses advanced AI technology to identify and localize microbiome samples to specific regions, bodies of water, and even individual cities.
The team's findings are striking: various locations boast unique bacterial populations. This means that everyday actions—like grabbing a handrail at a train station or bus stop—can transfer bacteria that provide a microbial fingerprint, potentially linking individuals back to their exact location.
Eran Elhaik, the lead researcher at Lund University, elaborated on this, stating, "Unlike human DNA, which remains relatively stable, the human microbiome is dynamic and changes with environmental interactions. By tracking where your microorganisms have recently resided, we can monitor disease spread, identify infection sources, and even aid in criminal investigations."
To grasp how the research team leverages bacteria to infer someone's recent activities, one must understand that microbial communities leave detectable geographical clues. The researchers developed their innovative approach by analyzing vast datasets of microbiome samples from diverse environments, which include urban settings, soil samples, and marine ecosystems. They trained an AI model to recognize the unique "fingerprints" of these microorganisms and correlate them with specific geographical locations.
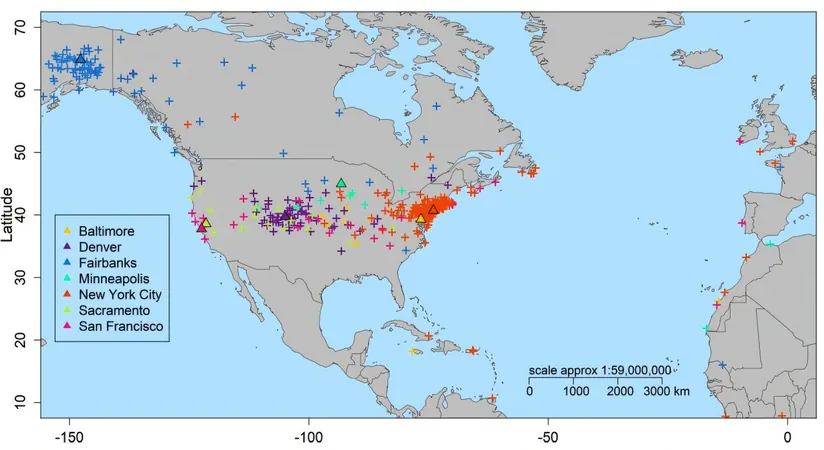
The sheer scale of their analysis was staggering: they sifted through 4,135 samples from public transport systems across 53 cities, 237 soil samples from 18 countries, and 131 marine samples from nine distinct water bodies. Remarkably, the team accurately pinpointed the city source for 92% of their urban samples.
In an impressive demonstration, the team traced samples collected in Hong Kong to their underground station of origin with an 82% accuracy rate. In New York City, the mGPS technology was even able to differentiate between the microbiomes found at a street kiosk and those on nearby handrails that were just a meter apart. As the volume of microbiome data continues to expand, Elhaik envisions that we are on the cusp of an entirely new era in forensic science.
"We are only scratching the surface of understanding the intricate relationship between microorganisms and their environments," Elhaik concluded. "Our next step is to map the microbiomes of entire cities. This endeavor could significantly enhance forensic investigations and deepen our understanding of the microbial ecosystems that share our lives—whether it’s in our streets, gardens, or even on our skin."
This revolutionary approach not only paves the way for more precise forensic analyses but also holds the promise of improving public health strategies by identifying and monitoring disease reservoirs in real-time. The implications of this research could be transformative, reshaping our understanding of microorganisms and their impact on human life and society.


 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)