Game-Changer in Dengue Virus Tracking: The New Naming System Revolutionizes Global Health
2024-09-30
Introduction
In a groundbreaking move poised to enhance global public health efforts, Yale researchers, along with scientists from 14 other countries, have devised an innovative naming system for dengue virus lineages. This systematic approach mirrors the successful genomic surveillance strategies that have been pivotal in monitoring the SARS-CoV-2 virus during the COVID-19 pandemic. With more than 12 million dengue cases reported this year alone, the new system arrives just in time to tackle this escalating health crisis effectively.
Dengue Fever Overview
Dengue fever, transmitted by mosquitoes, can cause debilitating symptoms such as high fever, severe headaches, body aches, nausea, and rashes, with severe cases potentially leading to death. While the virus predominates in tropical and subtropical regions, alarming outbreaks have surfaced in non-typical areas, including the United States and Italy. In 2023, the World Health Organization noted nearly 4.6 million cases in the Americas, marking a significant increase in infections from previous years.
Limitations of Traditional Methods
The traditional methods for categorizing dengue variants have proven insufficient, as different countries and research entities have adopted diverging systems, creating confusion and hindering effective tracking. Nathan Grubaugh, an associate professor at the Yale School of Public Health and senior author of the study, emphasized the urgent need for a unified approach, stating, "From a public health and vaccine development perspective, dengue virus genomic surveillance data is critically needed."
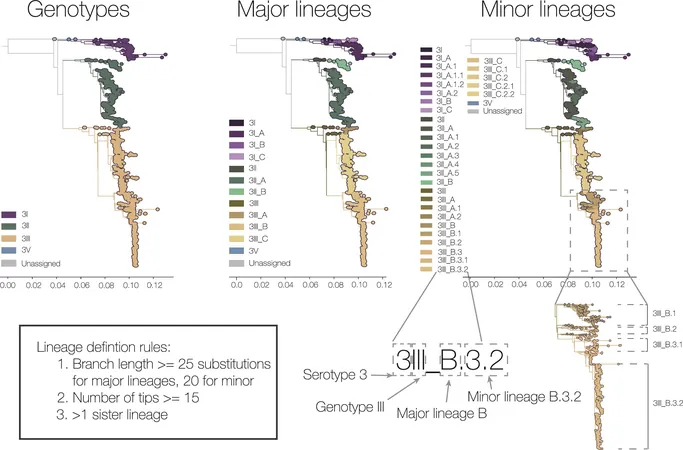
New Taxonomy for Dengue Virus
To address this challenge, Grubaugh and lead author Verity Hill collaborated with global colleagues to establish a cohesive framework for naming dengue virus lineages. This new taxonomy not only categorizes the virus into smaller, more manageable groups but also introduces two novel sub-genotype categories—major and minor lineages. This granular approach enables researchers to pinpoint and study the emergence of potentially dangerous variants more efficiently.
Advantages of the New System
Hill elaborated on the advantages of the new system: "With broad categories, it can appear as though regions are only impacted by one or two versions of the virus. However, increased resolution allows us to identify a multitude of circulating lineages." Key international experts, including co-author Anderson F. Brito from the All for Health Institute in São Paulo, Brazil, have hailed the system as an essential tool for monitoring the dengue virus.
Global Collaboration and Resources
In countries like Brazil, where dengue poses a serious public health risk, this system aids in recognizing emerging lineages and predicting outbreak impacts," Brito noted. He highlighted the value of the system in evaluating the effectiveness of new dengue vaccines recently introduced in Brazil. In addition to facilitating research, the team has launched a dedicated website offering access to the new naming system and resources for researchers worldwide.
Conclusion
This initiative includes an annual committee meeting where new lineages will be designated based on submitted genomic data, fostering a collaborative global research environment. This new system not only promises to revolutionize our understanding of the dengue virus but also holds the potential to significantly improve vaccine development and public health responses to outbreaks. With dengue's uncontrolled spread, the collaboration and rapid adaptation in scientific research showcased by these researchers may be the key to combatting this global health threat.

 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)