Exploring E. coli Genomes: The Intersection of Big Data and Biological Complexity
2024-11-08
Author: Siti
A Journey Through E. coli's History
First described in 1885, Escherichia coli (E. coli) stands as one of the most extensively studied organisms in the realm of microbiology, serving dual roles as a crucial commensal organism in the human gut and a notorious pathogen responsible for several outbreaks of foodborne illness. Today, researchers have analyzed over 10,000 E. coli genomes and discovered an eye-popping count of more than 100,000 distinct gene families within the organism's "pan-genome." As of October 2024, nearly a million E. coli genome sequences are cataloged in the NCBI database!
E. coli's Growing Genetic Library
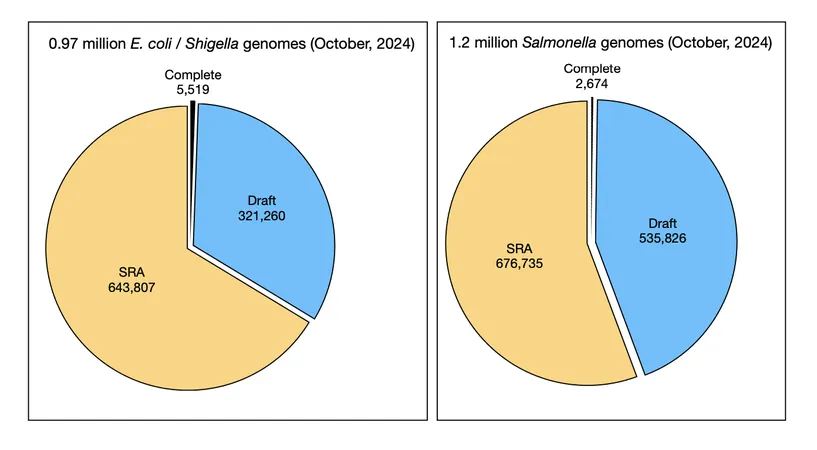
The pace of genomic sequencing is nothing short of astounding. Among the sequenced genomes are more than 5,000 complete E. coli and Shigella genomes alongside over 321,000 draft genomes. The Sequence Read Archive (SRA) doubles this number, indicating that the universe of E. coli genetic information is expanding rapidly. Notably, an average E. coli strain possesses around 5,000 genes, but the sheer volume of data leads to estimates of a staggering number of gene families across the microbial landscape.
Unlocking Insights in E. coli Genomics
The story of E. coli genomics is one of unraveling mysteries over time. The first genome, sequenced in 1997 from a laboratory strain, contained about 4,300 genes. This was merely the tip of the iceberg. Subsequent sequencing of pathogenic strains revealed additional genes not present in the laboratory variant, complicating our understanding. As more E. coli genomes were sequenced, methods evolved: microarray technology allowed for comparisons across various strains, and an extensive pan-genome was constructed detailing gene family variability.
The Future: Challenges and Opportunities in Big Data
The landscape of E. coli genomics is now interwoven with the complexities of big data. Researchers are tasked with managing vast, often messy datasets characterized by inconsistencies in organism naming and sequencing quality. For example, the Enterobacteriaceae family is well-researched, but predictions about future evolution remain uncertain. However, tools like the Mash program have emerged as reliable and scalable solutions to cluster genomes, demonstrating potential in managing massive datasets efficiently—a recent example includes clustering over a million Salmonella genomes into phylogroups in under a week.
Interestingly, the rapid pace of genomic naming has led to considerable controversy. Approximately 80% of bacterial species names have changed within the last two years as researchers propose new classifications based on genomic information. This also touched E. coli, prompting debates about the future handling of its nomenclature. The well-known laboratory strain E. coli K-12 was controversially renamed to align closer with Shigella flexneri, sparking resistance in the scientific community.
With the evolving field of microbial genomics, one thing is clear: the journey of E. coli—once a simplistic bacterium in textbooks—has evolved into a complex narrative filled with surprises and a future ripe for discovery. Stay tuned, as advancements in genetic research are poised to change our perception of this ubiquitous organism once again!



 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)