Disrupting the Double Helix: The Hidden World of Alternative DNA Structures in Ape Genomes
2025-04-24
Author: Ming
Unveiling Non-B DNA: A New Perspective on Ape Genomes
Have you ever wondered if DNA could be more than just the familiar double helix we all know? Recent research reveals that certain DNA sequences in great apes can form alternative structures known as non-B DNA. These unconventional shapes are turning out to be crucial players in cellular processes and the evolution of our genomes.
Groundbreaking Research from Penn State
Led by biologist Kateryna Makova at Penn State, a team has successfully predicted the locations of these elusive non-B DNA structures across great ape genomes. This game-changing research is paving the way to understanding how these configurations could influence genetic diseases and cancer.
Closing the Gaps: Advancements in Genomic Sequencing
The key to this groundbreaking study lies in the newly completed telomere-to-telomere (T2T) genomes of humans and their ape relatives. Until recently, many parts of these genomes remained a mystery, primarily due to the repetitive nature of non-B DNA that conventional sequencing struggled to decode. Thanks to advanced long-read sequencing technologies, researchers are finally filling in those gaps.
The Complexity of Assembly: A Jigsaw Puzzle of Genomes
Most genome sequencing approaches break DNA into tiny, manageable pieces—imagine a jigsaw puzzle with countless similar-looking pieces. Researchers can assemble these fragments into a coherent picture, but repetitive DNA complicates things significantly. With long-read technologies, scientists can tackle larger segments, making it easier to uncover the fascinating non-B DNA structures.
Diverse Forms of Non-B DNA: More Than Meets the Eye
Non-B DNA isn't just one shape; it comes in several intriguing forms like bent DNA, hairpins, G-quadruplexes, and Z-DNA. Each of these structures has been linked to crucial cellular functions such as DNA replication, gene regulation, and the stability of chromosome ends and centers—key players in cell division!
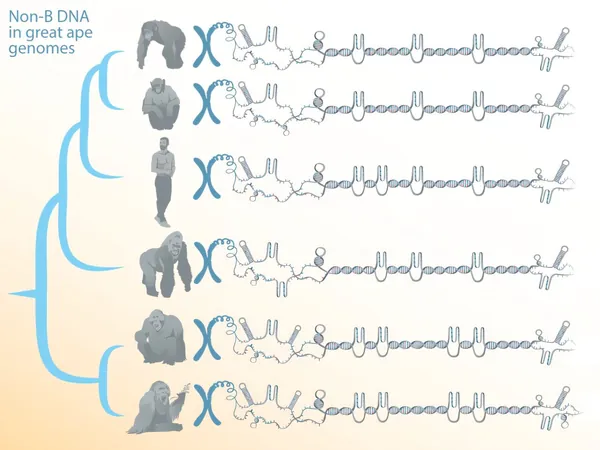
A Revolutionary Map of Ape Genomes
In their pursuit, the research team scoured T2T genomes for motifs that signal potential non-B DNA formation across humans, chimpanzees, bonobos, gorillas, orangutans, and even a lesser ape known as siamang. What they found was remarkable: a wealth of newly explored sequences rich in non-B motifs, demonstrating similarities across species.
Mutation Rates and Evolution: A Double-Edged Sword
Interestingly, non-B DNA is associated with higher mutation rates and chromosomal instability, which could lead to significant changes in genomes. For instance, in recent findings, non-B DNA structures were linked to a translocation on chromosome 21 associated with a form of Down Syndrome, hinting at a deeper, more complex relationship between these structures and genetic disorders.
The Next Frontier: Exploring the Context-Dependent Nature of Non-B DNA
While researchers are just beginning to confirm the presence of non-B DNA structures, they stress the importance of context. The formation of these structures may depend on various factors like cell type and developmental stage. This shift in scientific thinking moves us from viewing the genome merely as a sequence to understanding it as a functional and structural entity.
A Launchpad for Future Discoveries
Makova and her team aspire that this study will inspire further exploration into the intriguing roles of these non-B DNA structures in the genome. As we continue to delve into the rich tapestry of genetic information, the possibilities seem limitless.



 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)