Breakthrough Discovery: New Method Maps Hundreds of Proteins Inside Cell Nuclei – A Game Changer for Disease Understanding!
2024-12-18
Author: Siti
Introduction
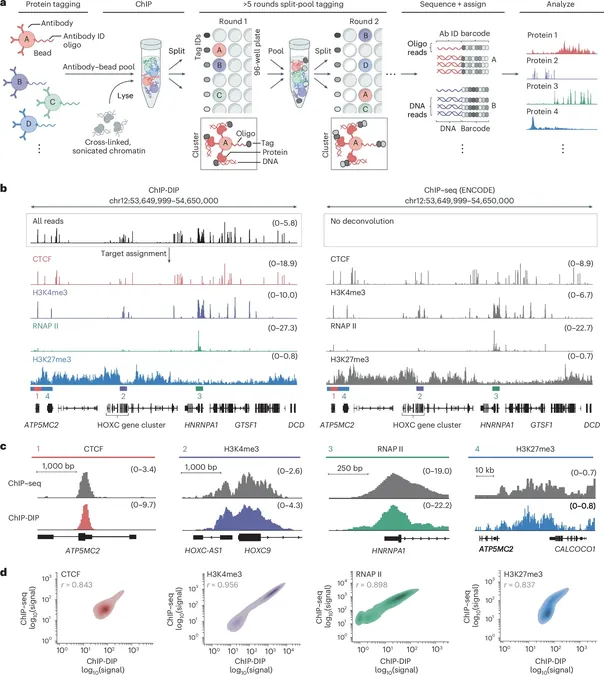
In a revolutionary development, researchers at Caltech have unveiled a groundbreaking method that allows scientists to map the positions of hundreds of DNA-associated proteins all at once within the cell nucleus.
Dubbed ChIP–DIP (Chromatin ImmunoPrecipitation Done In Parallel), this innovative technique is set to transform our understanding of gene regulation, especially in the contexts of disease and development. The research led by Professor Mitchell Guttman is detailed in the prestigious journal *Nature Genetics*.
Significance of the Discovery
What makes this discovery particularly exciting is the insight it provides into the human body's cells. Despite all cells containing the same DNA, they express different proteins that enable them to perform specialized functions. This diversity relies heavily on the regulation occurring within the nucleus, which houses thousands of regulatory proteins acting at specific locations.
To illustrate, consider that a cell’s DNA is a staggering two meters in length when fully extended, yet it must fit into a nucleus 50 times narrower than a human hair! This feat is achieved by winding the DNA around proteins called histones, which can be modified throughout a cell's life, exposing or obscuring various gene sections. Think of it as changing your wardrobe with the seasons—bringing forward certain items while storing others away.
Implications for Disease Understanding
The implications of this research are immense. When gene regulation fails, it can contribute to the onset of serious conditions such as cancer, neurodegenerative diseases, and inflammatory disorders. Until now, unraveling the mysteries of gene regulation has proven to be a monumental challenge; traditional methods could only analyze one protein at a time.
ChIP–DIP promises to change this by enabling the study of hundreds of DNA-associated regulatory proteins simultaneously, offering a rich, dynamic picture of the cellular landscape.
Quote from Research Team Members
As Isabel Goronzy, a former Ph.D. student in Guttman’s lab, explains, "Using ChIP–DIP, we’ve demonstrated that immune system cells rapidly modify their histone proteins following an inflammatory event, driving the activation of inflammatory genes." This has significant implications for understanding how our bodies respond to different stressors and injuries over time.
Adding to the excitement, ChIP–DIP has drastically accelerated research efforts—while previous international collaborations took nearly a decade for a few thousand experiments, this new method has completed over 500 experiments in just weeks! It provides incredible precision in mapping the intricate interplay of proteins that dictate gene activity in both healthy tissues and those impacted by disease.
Andrew Perez, a postdoctoral scholar and co-author of the study, highlights another advantage, saying, "With a single sample, we can map hundreds of proteins, allowing us to examine the epigenetic signatures—how environmental factors influence gene expression—of various diseases."
Conclusion
"This advancement has been a long-held aspiration within the genomics community," Guttman remarks. "ChIP–DIP represents a paradigm shift in our capabilities."
As we stand on the precipice of a new era in genomic research, this method could unlock the secrets to disease mechanisms and pave the way for novel therapies. Buckle up for what promises to be an electrifying journey into the very nucleus of life!


 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)