Breakthrough Discovery in Bacterial Antibiotic Resistance Unveils Potential New Therapies!
2025-01-23
Author: Jia
Overview
In a remarkable advancement for global public health, an international team of researchers has shed light on the intricate genetic mechanisms behind bacterial antibiotic resistance. This issue poses a significant challenge worldwide, as bacteria develop multiple defenses to withstand the effects of drugs designed to eliminate them.
Role of Plasmids in Resistance
One critical element in this resistance is the presence of plasmids—tiny DNA molecules found within bacteria that carry genetic information, including antibiotic resistance traits. Understanding the role these plasmids play is crucial; with this knowledge, scientists can formulate innovative therapeutic strategies aimed at combatting drug-resistant infections.
Research Collaboration
Researchers from the John Innes Center, in collaboration with specialists from Madrid, New York, and Birmingham, explored a model plasmid known as RK2. This plasmid is widely used in research to understand how resistance is transmitted in clinical settings. Their findings were published in the esteemed journal *Nature Microbiology* under the study titled, "KorB switching from DNA-sliding clamp to repressor mediates long-range gene silencing in a multi-drug resistance plasmid."
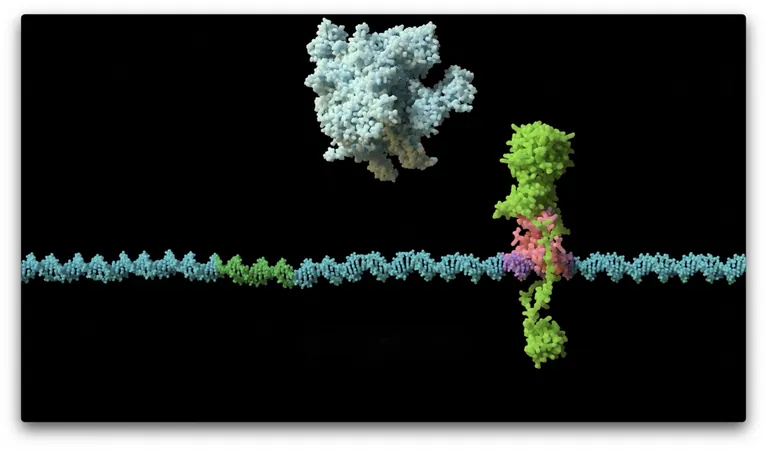
Discovery of KorB and KorA
The focal point of the study was a protein named KorB, essential for the survival of plasmids within their bacterial hosts. While previously recognized for its role in regulating gene expression, the precise mechanics remained elusive until now. By leveraging advanced microscopy and protein crystallography techniques, the team discovered a fascinating interaction between KorB and another molecule known as KorA.
Mechanism of Gene Regulation
This KorB-KorA regulatory system operates like a pair of complementary components—KorB acts as a sliding clamp while KorA serves as a lock, immobilizing KorB at strategic locations within the DNA. This coordinated interaction effectively shuts down gene expression, allowing the plasmid to persist securely within the bacterial cell.
Implications for Future Research
The revelation of this mechanism not only enhances our understanding of long-range gene regulation in bacteria but also opens the door for future therapeutic innovations. As Dr. Thomas McLean, the study's lead author and a postdoctoral researcher at the John Innes Center, aptly points out, this breakthrough was a product of "curiosity-driven science." An unexpected outcome from what was intended as a routine experiment on a Friday afternoon pivoted their research towards the key interactions governing plasmid stability.
Significance of the Findings
This research marks a significant leap forward in addressing a question that has puzzled scientists for decades: How does the essential protein KorB dictate when genes are activated or silenced in the multi-drug-resistant RK2 plasmid?
Conclusion and Future Directions
The implications of this study are vast, as the research team aims to expand their analysis to include more clinically relevant plasmids. By delving deeper into the KorB-KorA mechanism, they hope to unravel how this system disassembles at appropriate times, ultimately paving the way for groundbreaking therapeutics that could destabilize plasmids and enhance the effectiveness of existing antibiotics.
Stay tuned as this exciting research unfolds, potentially redefining our approach to tackling one of humanity's most pressing health issues!

 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)