Breakthrough Discovery: Eight New Enzymes Unleashed for Faster Cancer Drug Production!
2025-06-16
Author: Nur
In a groundbreaking revelation, researchers at Stanford University have uncovered eight entirely new genes that could revolutionize the production of the critical cancer drug, Taxol. By expressing these genes in tobacco leaves, the team successfully replicated the Taxol precursor, baccatin III, at levels equal to that found in yew needles. This discovery could significantly accelerate the process of manufacturing this life-saving medication.
The Challenge of Extracting Taxol from Nature
Yew trees (Taxus species) are the natural source of paclitaxel, the active ingredient in Taxol, a potent chemotherapy drug. However, these slow-growing trees only yield tiny amounts of crucial compounds—baccatin III makes up a meager 0.001-0.050% of their dry weight, predominantly found in the bark. The extraction process is so inefficient that manufacturers often rely on a semi-synthetic method, converting baccatin III into paclitaxel.
Ecological and Economic Costs of Current Methods
Harvesting yew bark not only jeopardizes the trees’ survival but also carries high ecological and economic costs. While needle collection is a more sustainable approach, it yields lower concentrations of the desired compounds. Therefore, a fully sustainable biosynthetic process is urgently needed.
Decoding the Enzymatic Mystery
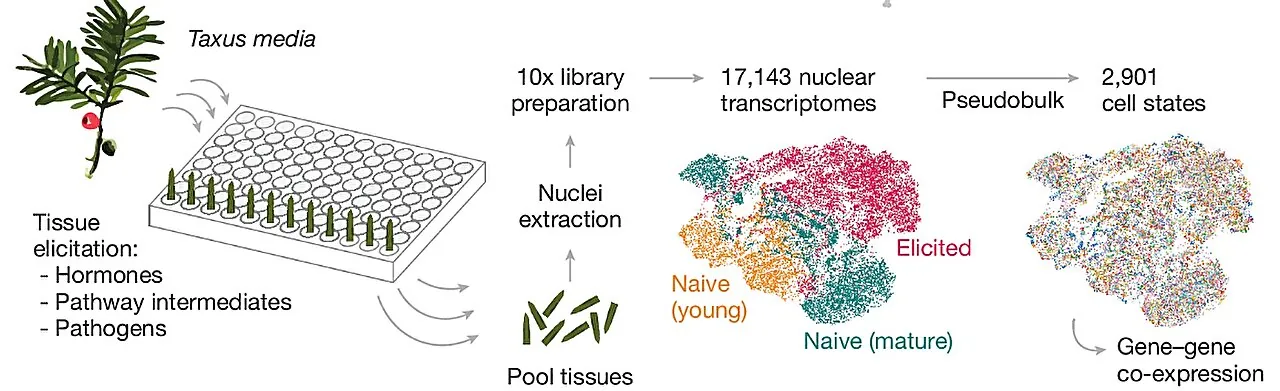
From the late 1990s until 2006, researchers identified 12 key enzymes in the Taxol synthesis pathway, but progress stalled, leaving numerous steps unexplored. Recent research titled "Discovery of FoTO1 and Taxol genes enables biosynthesis of baccatin III," published in Nature, utilized innovative multiplexed perturbation × single-nucleus (mpXsn) transcriptomics to analyze Taxus metabolic pathways comprehensively.
Cutting-Edge Technique: mpXsn Transcriptomics
By examining a staggering 17,143 single-nucleus transcriptomes derived from a variety of tissue samples and conditions, the researchers created a condensed dataset of 2,901 pseudo-bulk cell states. They then tested top candidates through transient expression in tobacco leaves, analyzing the metabolic products with advanced techniques like gas chromatography–mass spectrometry.
Game-Changing Enzymes Discovered
Emerging from these analyses were eight novel genes, including FoTO1, a protein that enhances the accuracy of the critical first oxidation step in the pathway. Other newly discovered enzymes were pivotal in converting substrates into baccatin III. Interestingly, two of these enzymes, despite evolving independently, fulfilled a similar catalytic role.
A Major Leap Forward for Drug Manufacturing
The authors of the study note that the unique identities of these new genes explain their historical obscurity, revealing the necessity for unexpected gene families and functions in pathway reconstitution. The mpXsn method not only broke through transcriptomic barriers within complex genomes but also provided a clearer view of the natural processes behind paclitaxel synthesis.
Toward Sustainable Large-Scale Production
These newly identified enzymes could serve as a sustainable alternative to reliance on yew trees for baccatin III, paving the way for eco-friendly, large-scale production of paclitaxel. Furthermore, the mpXsn platform holds promise for studying gene functions in other challenging organisms, particularly eukaryotes, which traditionally face hurdles in functional genomics due to the lack of complete gene clusters found in prokaryotes.
The Future of Functional Genomics
With the mpXsn method enabling more affordable and precise tracking of gene covariance across numerous transcriptional states, it may finally address long-standing obstacles in functional genomics, opening doors for significant scientific advancements.
 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)