Breakthrough AI Model Set to Revolutionize Genomic Predictions for Disease Treatment
2024-11-11
Author: Jia
Breakthrough AI Model Set to Revolutionize Genomic Predictions for Disease Treatment
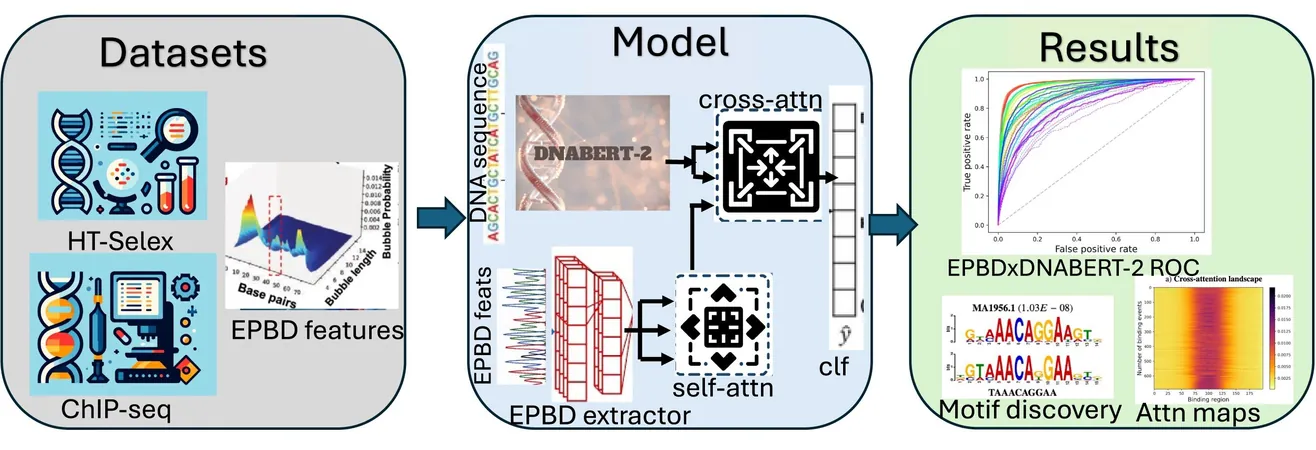
In a groundbreaking advancement at Los Alamos National Laboratory, scientists have unveiled the first multimodal deep learning model specifically designed to decode the complex interactions between DNA and disease. Named EPBDxDNABERT-2, this innovative AI model harnesses the unique phenomenon known as DNA breathing—the natural opening and closing of the DNA double helix—to better predict how transcription factors, the proteins that control gene activities, bind to the human genome.
Los Alamos researcher Anowarul Kabir, the study's lead author published in Nucleic Acids Research, emphasized the enormity of the challenge: "With a human genome that contains 3 billion base pairs, determining which transcription factors bind to specific DNA locations is crucial for understanding gene regulation associated with various diseases."
Decoding DNA Through AI
DNA serves as the essential blueprint for human existence, and its regulation is key to many diseases including cancer. Transcription factors play a vital role in gene expression, and accurately predicting their binding sites can illuminate pathways for drug development and personalized medicine.
The researchers began by training their foundational model on extensive DNA sequence data. They then developed a sophisticated DNA simulation program that accounts for the dynamics of DNA interactions. The combined effort led to the creation of EPBDxDNABERT-2, allowing the processing of genomic sequences across entire chromosomes while feeding in valuable DNA dynamics data.
The Power of DNA Breathing
Central to the model's success is the concept of DNA breathing. This process is closely linked to transcriptional activity, as the spontaneous opening of the DNA strand correlates with how well transcription factors can access and regulate genes. "By integrating DNA breathing features into our model architecture, we significantly enhanced the accuracy of predicting transcription factor binding," stated researcher Manish Bhattarai.
The investigators employed the laboratory's advanced supercomputer, Venado, which marries CPU and GPU technologies to optimize AI tasks. This powerful computational resource allowed them to analyze expansive datasets, utilizing gene sequencing information from 690 experimental results involving 161 distinct transcription factors across 91 human cell lines. Remarkably, EPBDxDNABERT-2 increased prediction accuracy by an impressive 9.6% for binding over 660 transcription factors.
A Tool for Future Discoveries
While DNA breathing alone can yield nearly accurate assessments of transcriptional activity, this new multimodal model extracts specific binding motifs—crucial sequences where transcription factors attach. The versatility and effectiveness of EPBDxDNABERT-2 across varied datasets signals a promising future for computational genomics.
"This tool not only enhances our understanding of complex biological processes but offers an advanced method for exploring gene regulation mechanisms directly tied to disease treatment strategies," declared Bhattarai.
In a world where rapid advancements in AI technology intersect with critical fields like genomics, EPBDxDNABERT-2 stands at the forefront of a new era in personalized medicine, heralding a future where diseases can be tackled at their genetic roots. Scientists anticipate that this revolutionary model will pave the way for the development of targeted therapeutics, ultimately transforming healthcare as we know it.



 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)