Breakthrough AI from Microsoft Can Predict Protein Movement in Minutes!
2025-07-10
Author: John Tan
Unlocking Protein Secrets with AI Technology
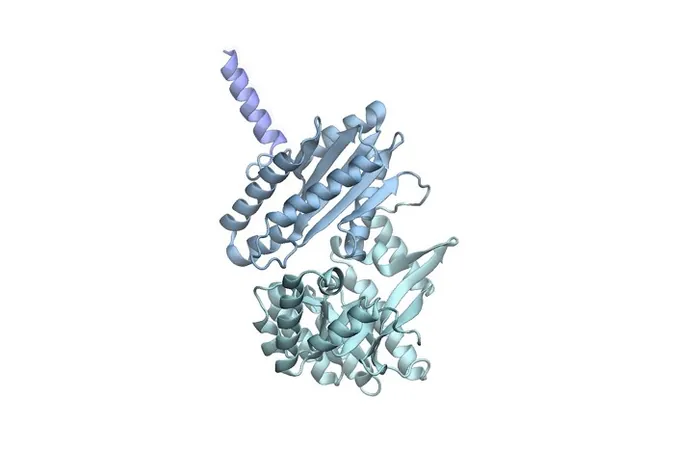
Proteins are the workhorses of biology, dynamically twisting and turning to perform vital functions. Thanks to a groundbreaking AI tool by Microsoft, known as BioEmu, researchers can now predict the myriad ways proteins can move in just minutes—using a fraction of the resources traditionally required.
The Revolution in Protein Structure Understanding
Led by Frank Noé at Microsoft Research’s AI for Science lab, BioEmu stands out as a game changer in the field. Building on the foundation laid by pioneering tools like AlphaFold—a tool that revealed around 200 million static protein structures—BioEmu goes a step further by revealing the dynamic movements proteins can make.
Historically, scientists relied on lengthy and resource-intensive methods such as cryogenic electron microscopy and molecular dynamics simulations to decipher protein behavior. But these methods are often time-consuming and expensive. Enter BioEmu, a deep learning tool designed to emulate protein movement rather than simulate it, drastically speeding up the process.
How BioEmu Works: A Deep Learning Marvel
BioEmu is based on a sophisticated deep learning neural network trained on a blend of costly experimental and historical simulated data. Noé explains that the tool predicts not just how proteins will move, but also the structures they can stabilize, delivering results with an accuracy of 1 kcal/mol—an achievement he terms 'experimental accuracy.'
The Drug Discovery Potential
One of the most exciting applications for BioEmu lies in drug discovery. With its ability to identify hidden binding pockets in proteins, researchers may pinpoint new ways to inhibit harmful proteins that drive diseases. While it may take time to translate this potential into tangible medical breakthroughs, the promise is undeniable.
Praise from Experts and Future Prospects
Experts are already singing BioEmu’s praises. Zhidian Zhang, a postdoc at MIT, highlights its unique ability to predict the distribution of protein conformations, a complex challenge that existing tools struggle to address. Alberto Perez from the University of Florida, eager to integrate BioEmu into his work, applauds its open-source release, noting the prior disappointment when AlphaFold 3 withheld its code.
Addressing Limitations and Looking Ahead
However, not all is perfect; BioEmu has its limitations. It performs best with smaller proteins and faces challenges with larger ones and proteins interacting with ligands or membranes, both critical areas in drug development. Moreover, it doesn't account for the kinetics of protein movements, which is essential for understanding the timing of these transformations.
Despite its current restrictions, both Zhang and Perez, along with Noé, are optimistic that future iterations of BioEmu will enhance its capabilities. Perez recalls the gradual evolution of AlphaFold, suggesting that just like its predecessors, BioEmu could soon be a transformative force in the field.



 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)