Revealing the Secrets of tRNA Cleavage: A Breakthrough in Protein Synthesis!
2025-07-07
Author: Jacques
In the intricate world of cellular biology, transfer RNA (tRNA) holds a pivotal role as a key player in protein synthesis. Acting as couriers, tRNAs translate genetic instructions from messenger RNA (mRNA), delivering essential amino acids to ribosomes—the cell’s protein factories. However, before tRNAs can spring into action, they must undergo precise trimming and shaping.
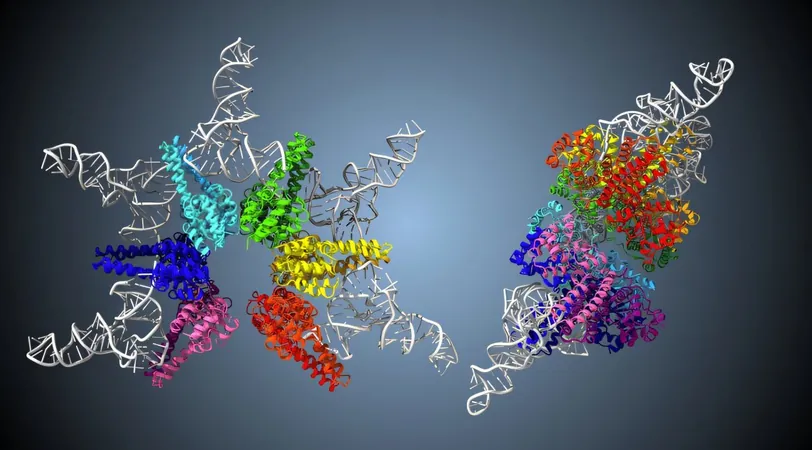
Now, a groundbreaking study from researchers at Kyushu University has unveiled the workings of HARP, the smallest known enzyme responsible for tRNA processing. This star-shaped complex comprises 12 protein molecules, enabling it to make dual cuts at both the 5' and 3' ends of tRNA. The researchers aim for their discoveries to significantly advance the fields of synthetic biology and biotechnology, ultimately leading to innovative tools and artificial enzymes.
Understanding the Enzymatic Process: A Closer Look at HARP
In biological systems, proteins require meticulous processing to function effectively. tRNAs, in particular, undergo various transformations, including the trimming of their ends—the 5' and 3' ends. The enzyme RNase P plays a crucial role in this cutting process, existing in two major forms: one predominantly made of RNA and another entirely composed of proteins. The RNA-based RNase P has been extensively studied, while its protein counterpart, which includes variants like PRORP and HARP, remains less understood.
HARP, short for Homologs of Aquifex RNase P36, boasts a unique six-pointed star shape and is notably compact, found in select bacteria and archaea. Despite its intriguing structure, the complexities of its function and form were previously shrouded in mystery.
Cutting-Edge Discoveries: Visualization and Functionality
To demystify how HARP interacts with pre-tRNA, researchers employed cryogenic electron microscopy (cryo-EM) for advanced imaging. Their findings revealed a radial structure where pre-tRNA molecules bind to five specific sites on the HARP enzyme. This setup transforms HARP into a "molecular ruler," allowing it to accurately gauge distances from the tRNA's 5' end to a critical cleavage point.
Even more astonishing, this method of precise measurement was observed in other forms of RNase P, hinting at a fascinating example of convergent evolution across various life forms.
Unveiling New Dimensions of tRNA Processing
The research further uncovered a surprising detail: HARP binds only five pre-tRNA molecules, contrary to earlier predictions of ten, leaving seven active sites vacant. This led to an intriguing second discovery: when the researchers performed cleavage assays, they found a product corresponding to the pre-tRNA's 3' end. This revelation indicates that HARP first trims the 5' end and then utilizes its remaining sites for 3' end cleavage.
As Professor Yoshimitsu Kakuta explains, the oligomerization of the small protein HARP endows it with dual functionality in tRNA processing, showcasing an evolutionary strategy that enables organisms with compact genomes to develop multifunctional enzymes.
A Bright Future for Synthetic Biology and Biotechnology!
The implications of these findings extend far beyond basic science. By unraveling how flexible arrangements of structural elements can yield new functionalities, researchers lay the groundwork for future advancements in synthetic biology and biotechnological applications. Clearly, understanding the secrets of tRNA processing can spark a revolutionary change in how we approach biological engineering!








 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)