Groundbreaking Tool Reveals Hidden Protein Variants Linked to Genetic Mutations
2025-06-16
Author: Jacob
Unlocking the Secrets of Proteins
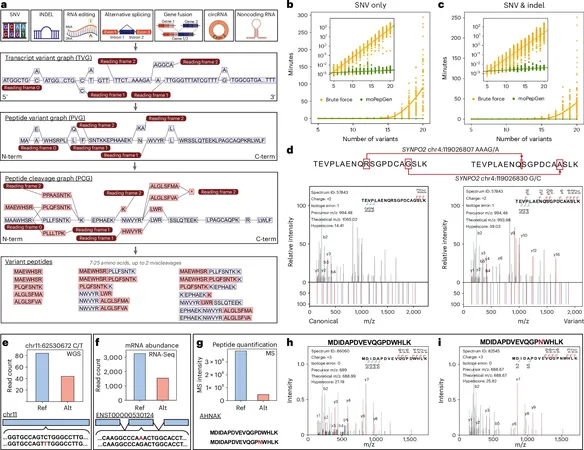
In a phenomenal advancement for cancer research and genetic science, scientists from UCLA and the University of Toronto have unveiled **moPepGen**, a cutting-edge computational tool designed to identify elusive genetic mutations in proteins. This breakthrough could redefine our understanding of diseases like cancer and neurodegenerative disorders.
Bridging the Gap Between Genetics and Disease
Published in **Nature Biotechnology**, moPepGen offers a revolutionary way to trace how DNA changes affect protein structures, opening doors to new diagnostic tests and treatment targets that have remained shrouded in mystery until now. This tool is a game-changer in the field of **proteogenomics**, which integrates genomics and proteomics to deliver a complete molecular profile of various diseases.
Confronting Detection Challenges
One of the major hurdles in this area has been the difficulty in accurately detecting variant peptides, which has hampered the identification of protein-level genetic mutations. Existing tools often fall short in capturing the full spectrum of protein variations. Enter moPepGen—a solution designed to tackle this precise challenge.
"We developed moPepGen to help researchers determine which genetic variants are actively expressed at the protein level, addressing a long-standing issue in our field,” said **Chenghao Zhu, Ph.D.**, a postdoctoral scholar at UCLA and co-first author of the study.
A Leap in Precision
This innovative tool employs a **graph-based approach** to analyze all types of genetic alterations, significantly enhancing the detection of hidden protein variations. As Zhu explains, "This allows us to see the full diversity of proteins and understand how mutations contribute to diseases more accurately." Proteins are vital in nearly all biological functions, and their structural changes can indicate disease progression, particularly in cancers.
Beyond Basic Changes
Unlike conventional methods that primarily note straightforward genetic changes like single amino acid shifts, moPepGen delves deeper, identifying a broad range of protein variations from alternative splicing to gene fusions. It systematically models the expression of genes into proteins, greatly increasing the detection capacity for mutations associated with diseases.
A New Era of Cancer Research
To showcase its prowess, the moPepGen team analyzed proteogenomic data from a variety of sources, including five prostate tumors and eight kidney tumors, uncovering previously hidden protein variations linked to genetic mutations and molecular changes. Remarkably, they detected **four times more** unique protein variants compared to traditional methods.
One of the most thrilling prospects for moPepGen lies in the realm of immunotherapy. It can identify cancer-specific variant peptides that might act as neoantigens, essential for developing personalized cancer vaccines and targeted therapies.
Accessible and Impactful
“By simplifying the analysis of complex protein variations, moPepGen has the potential to transform research across cancer, neurodegenerative diseases, and beyond,” states **Paul Boutros, Ph.D.**, co-senior author of the study and a prominent figure in precision medicine at UCLA.
The tool is available for researchers globally and can seamlessly integrate into existing proteomics workflows, ensuring its accessibility.
A Bright Future Ahead
With moPepGen at the forefront, scientists are now better equipped to bridge the gap between genetic data and real-world protein expression, heralding a new era in precision medicine and paving the way for groundbreaking treatments.







 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)