Breakthrough Study Reveals Simple Mathematics Behind Protein Mutation Effects – A Game Changer for Disease Treatment and Biotechnology!
2024-09-25
Author: Amelia
Introduction
In a groundbreaking study published in the esteemed journal *Nature*, researchers from the Center for Genomic Regulation (CRG) and the Wellcome Sanger Institute have unveiled a striking finding: mutations that affect protein stability adhere to remarkably straightforward mathematical rules. This discovery could expedite the development of new treatments for various diseases and enhance the design of proteins for industrial purposes.
The Role of Proteins and Mutations
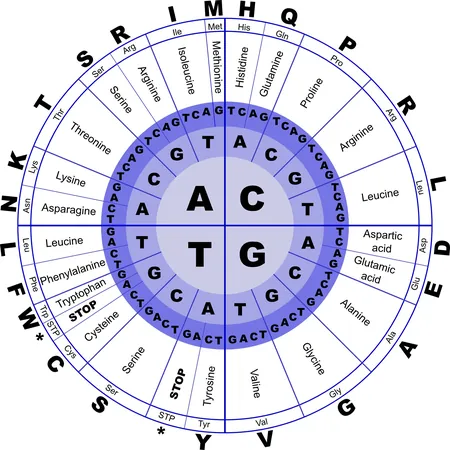
Proteins, the essential workers in biological systems, are composed of chains of twenty different types of amino acids. When a single amino acid is mutated, it can alter the entire shape of the protein, which can mean the difference between health and disease. Many genetic disorders, from cancer to neurodegenerative diseases, are linked to multiple mutations within a protein.
The Challenge of Mutation Combinations
Understanding how mutations change protein structure is crucial for assessing their role in disease. However, given the potentially astronomical number of combinations of mutations in proteins, thorough experimental testing has long been considered impractical. A protein made up of just 34 amino acids can have 17 billion different mutation combinations for a single amino acid change—a feat that would take 539 years to test if each combination required a second of testing.
Research Findings
As protein length increases, the number of possible combinations escalates dramatically. For instance, for a 100-amino-acid-long protein, the number of potential combinations surpasses the total number of atoms in the observable universe. Most known proteins, particularly those associated with human diseases, are considerably longer.
However, research led by Dr. André Faure at CRG suggests that the landscape of protein mutations is more navigable than previously understood. Historically, the scientific community was wary of the unexpected interactions between mutations, fearing they might create unpredictable shifts in protein structure.
"Contrary to longstanding beliefs, our research shows that while mutations can interact, such occurrences are relatively uncommon. Most mutations influence a protein's stability independently, allowing us to simplify our predictive models enormously," explains Aina Martí Aranda, a co-author of the study.
Implications for Disease Treatment and Biotechnology
Remarkably, the researchers generated thousands of protein variants, experimenting with various combinations of mutations to assess their stability. Their findings demonstrated that the overall effects of these mutations could often be calculated by simply summing the individual effects—a method that significantly reduces the complexity traditionally associated with protein mutation studies.
This new understanding holds substantial promise for improving the treatment of genetic diseases, where patients often present with a variety of mutation combinations affecting a single protein. Clinicians could leverage these insights to predict how different mutations impact protein stability and overall disease severity, leading to tailored treatment approaches that enhance patient outcomes.
Moreover, the implications extend to the realm of drug development. With clearer insights into which mutations destabilize proteins, researchers can now design intervention strategies for conditions such as Alzheimer's disease, where misfolded proteins like amyloid-beta lead to plaque formation in the brain.
The study opens new avenues for biotechnologists working on innovative solutions, such as designing enzymes capable of breaking down environmental plastics. By strategically introducing beneficial mutations, it is possible to engineer enzymes with improved activity and durability.
Limitations and Future Research
Nonetheless, the researchers acknowledge limitations in their findings, including the complexity of mutations involving three or more interactions. These higher-order interactions could significantly influence protein stability and are not always captured by their simplified models.
Despite the exciting potential, both experimental validation and further research are necessary, especially concerning applications in drug development, where unforeseen effects may arise. Nevertheless, this paradigm shift unveils a new chapter in our understanding of protein mutations, promising advancements in medicine and biotechnology that could change lives.
Conclusion
Stay tuned as this research paves the way for revolutionary treatments and groundbreaking innovations in the field!








 Brasil (PT)
Brasil (PT)
 Canada (EN)
Canada (EN)
 Chile (ES)
Chile (ES)
 Česko (CS)
Česko (CS)
 대한민국 (KO)
대한민국 (KO)
 España (ES)
España (ES)
 France (FR)
France (FR)
 Hong Kong (EN)
Hong Kong (EN)
 Italia (IT)
Italia (IT)
 日本 (JA)
日本 (JA)
 Magyarország (HU)
Magyarország (HU)
 Norge (NO)
Norge (NO)
 Polska (PL)
Polska (PL)
 Schweiz (DE)
Schweiz (DE)
 Singapore (EN)
Singapore (EN)
 Sverige (SV)
Sverige (SV)
 Suomi (FI)
Suomi (FI)
 Türkiye (TR)
Türkiye (TR)
 الإمارات العربية المتحدة (AR)
الإمارات العربية المتحدة (AR)